lactate dehydrogenase Ba
ZFIN 
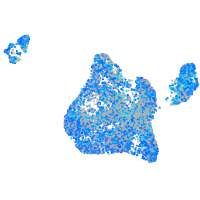
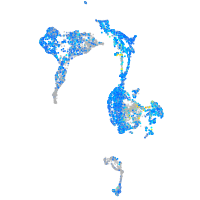
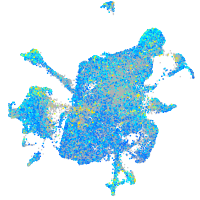
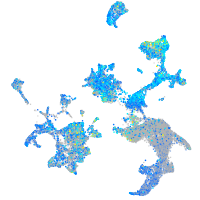
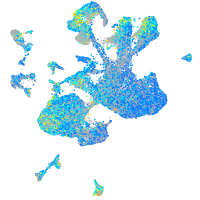
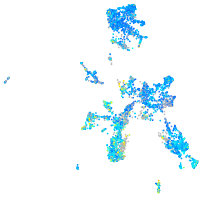
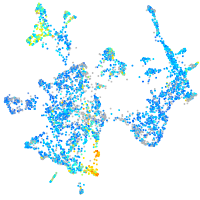
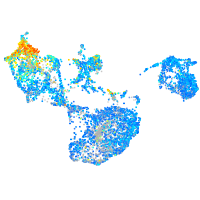
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dap1b | 0.490 | stmn1a | -0.560 |
| mdh2 | 0.481 | tpx2 | -0.558 |
| atp6v1e1b | 0.477 | gra | -0.555 |
| ppiab | 0.475 | nasp | -0.539 |
| gapdhs | 0.470 | ddx4 | -0.514 |
| rpl37 | 0.469 | hnrnpub | -0.514 |
| rps10 | 0.459 | syncrip | -0.510 |
| h3f3a | 0.457 | hnrnpd | -0.505 |
| zgc:114188 | 0.451 | rpl7l1 | -0.498 |
| b2ml | 0.448 | ebna1bp2 | -0.465 |
| rnasekb | 0.448 | NC-002333.4 | -0.464 |
| ccni | 0.446 | srsf5a | -0.462 |
| ndrg1a | 0.444 | dnd1 | -0.457 |
| rps27.1 | 0.441 | seta | -0.457 |
| rps17 | 0.438 | sf3b4 | -0.453 |
| zgc:123010 | 0.437 | akap12b | -0.449 |
| tmem258 | 0.429 | hnrnpabb | -0.448 |
| vdac3 | 0.428 | ppig | -0.446 |
| neo1b | 0.425 | sfpq | -0.445 |
| zgc:56493 | 0.417 | ranbp1 | -0.444 |
| eef1da | 0.413 | pttg1 | -0.444 |
| uqcc3 | 0.413 | chaf1a | -0.441 |
| slc25a5 | 0.412 | tubb2b | -0.440 |
| srd5a2a | 0.409 | ddit4 | -0.435 |
| rpl12 | 0.409 | kri1 | -0.428 |
| COX7A2 (1 of many) | 0.405 | nkap | -0.428 |
| si:dkey-16p21.8 | 0.403 | rrp15 | -0.426 |
| ndufa3 | 0.403 | stm | -0.423 |
| samd13 | 0.403 | hnrnpa0b | -0.419 |
| anxa4 | 0.402 | dynll2a | -0.418 |
| rps14 | 0.402 | safb | -0.417 |
| agrn | 0.402 | apoeb | -0.413 |
| tex264a | 0.400 | pdap1a | -0.412 |
| pcbp2 | 0.398 | ppan | -0.410 |
| cox6a1 | 0.398 | gnl3 | -0.409 |