La ribonucleoprotein 4Ab
ZFIN 
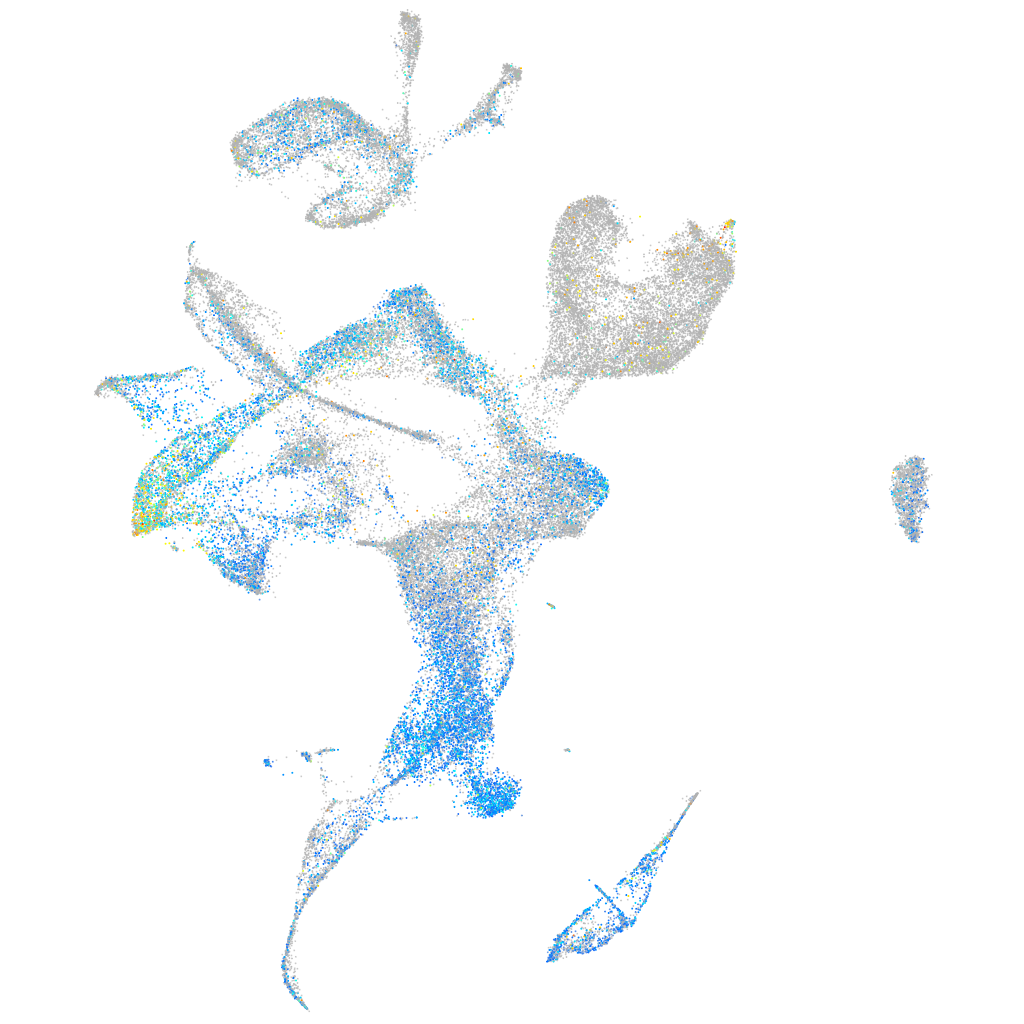
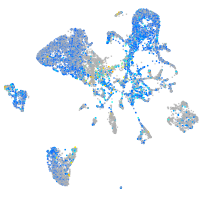
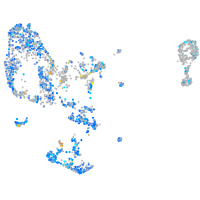
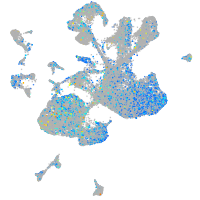
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gnat2 | 0.253 | gpm6aa | -0.217 |
| gnb3b | 0.248 | ptmaa | -0.193 |
| pdcb | 0.247 | marcksl1b | -0.178 |
| prph2a | 0.246 | mdkb | -0.176 |
| pde6c | 0.243 | nova2 | -0.172 |
| zgc:73359 | 0.243 | ndrg4 | -0.170 |
| grk7a | 0.239 | gpm6ab | -0.168 |
| rcvrn2 | 0.238 | vsx1 | -0.161 |
| cnga3a | 0.238 | scrt2 | -0.154 |
| slc25a3a | 0.237 | hmgb1a | -0.153 |
| ckmt2a | 0.236 | marcksl1a | -0.153 |
| chchd10 | 0.232 | ptmab | -0.151 |
| guk1b | 0.231 | calb2b | -0.149 |
| grk1b | 0.231 | gng13b | -0.148 |
| prph2b | 0.228 | ubc | -0.136 |
| gngt2b | 0.228 | rtn1a | -0.136 |
| rcvrn3 | 0.227 | cspg5a | -0.135 |
| rbp4l | 0.225 | gnb3a | -0.133 |
| si:ch1073-469d17.2 | 0.222 | pcbp3 | -0.132 |
| si:ch211-207l14.1 | 0.222 | pcp4l1 | -0.129 |
| cngb3.2 | 0.221 | btg1 | -0.124 |
| rgs9a | 0.220 | neurod4 | -0.123 |
| arl3l2 | 0.219 | isl1 | -0.123 |
| hexb | 0.218 | dscaml1 | -0.123 |
| si:ch211-81a5.8 | 0.217 | tmem178b | -0.121 |
| si:ch1073-303d10.1 | 0.214 | tmsb4x | -0.120 |
| inaa | 0.213 | zc4h2 | -0.120 |
| arl3l1 | 0.212 | ccng2 | -0.119 |
| gucy2d | 0.212 | myt1b | -0.119 |
| guca1c | 0.212 | mpp6b | -0.117 |
| c1qbp | 0.211 | lin7a | -0.116 |
| si:dkey-17e16.15 | 0.210 | gnao1b | -0.114 |
| eno1b | 0.208 | olfm1b | -0.113 |
| rcvrna | 0.207 | gng3 | -0.113 |
| ncaldb | 0.206 | ip6k2b | -0.112 |