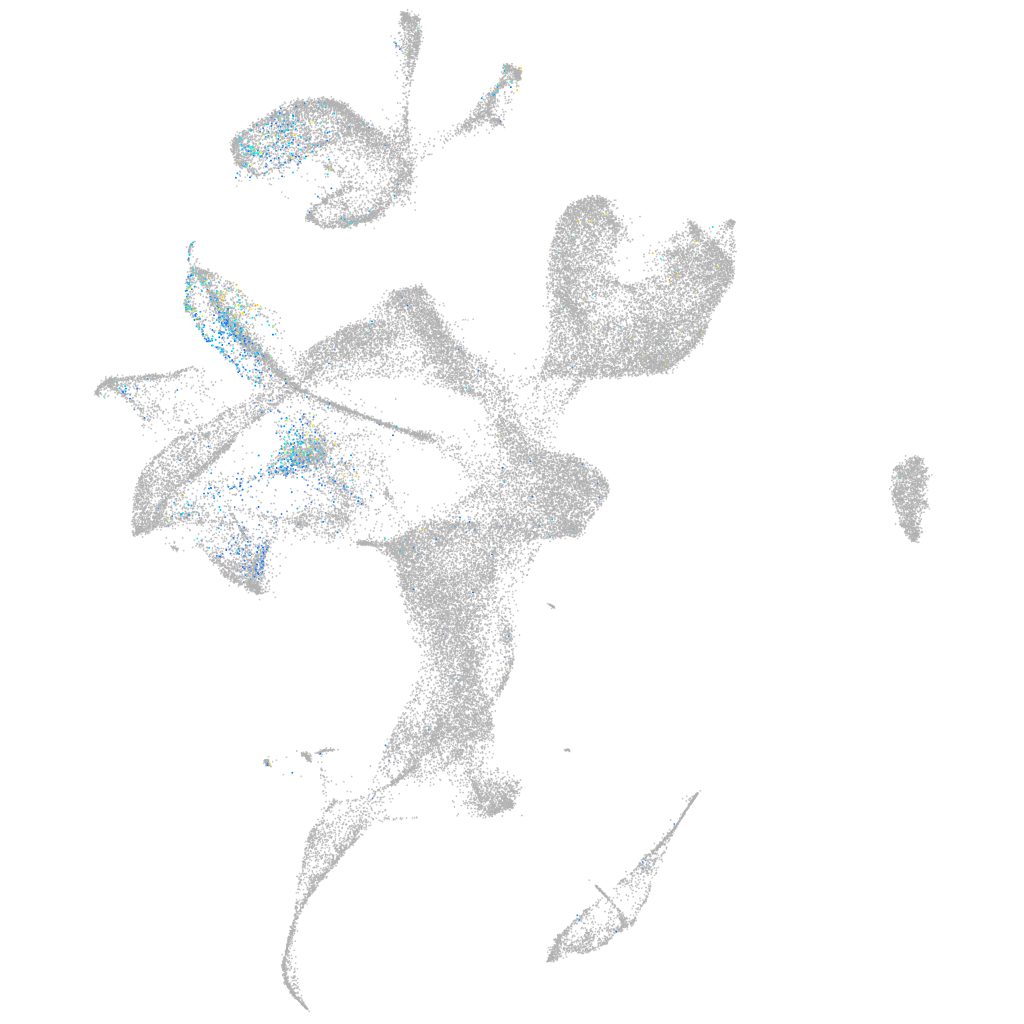
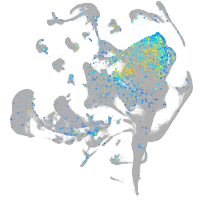
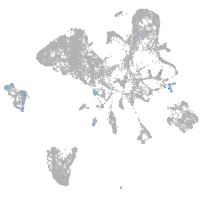
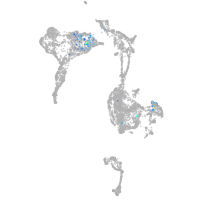
"L1 cell adhesion molecule, paralog a"
ZFIN 
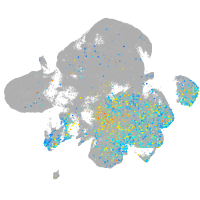
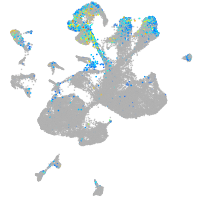
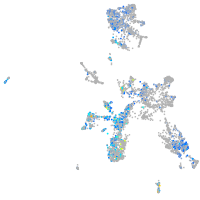
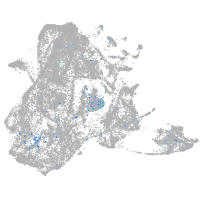
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ribc1 | 0.268 | hmgb2a | -0.125 |
| stmn2a | 0.256 | hmga1a | -0.072 |
| elavl4 | 0.238 | ahcy | -0.071 |
| islr2 | 0.237 | pcna | -0.068 |
| rtn1b | 0.232 | rrm1 | -0.065 |
| maptb | 0.229 | stmn1a | -0.064 |
| xpr1a | 0.227 | ccnd1 | -0.064 |
| gap43 | 0.226 | mdka | -0.062 |
| zgc:65894 | 0.220 | chaf1a | -0.062 |
| tmsb2 | 0.201 | dut | -0.060 |
| ywhag2 | 0.200 | nutf2l | -0.060 |
| snap25a | 0.196 | msi1 | -0.060 |
| stmn4 | 0.195 | mcm7 | -0.059 |
| rbpms2a | 0.191 | mki67 | -0.059 |
| nrn1a | 0.187 | lbr | -0.058 |
| celf4 | 0.187 | fabp7a | -0.058 |
| stmn2b | 0.186 | hmgb2b | -0.058 |
| id4 | 0.186 | selenoh | -0.057 |
| uchl1 | 0.185 | ccna2 | -0.057 |
| sv2a | 0.184 | msna | -0.057 |
| slc17a6b | 0.183 | dek | -0.055 |
| gng3 | 0.181 | tuba8l | -0.055 |
| inab | 0.180 | banf1 | -0.055 |
| tuba2 | 0.180 | neurod4 | -0.055 |
| tubb5 | 0.180 | lig1 | -0.054 |
| gabrb4 | 0.180 | cks1b | -0.054 |
| eno2 | 0.179 | mcm6 | -0.053 |
| zfhx3 | 0.179 | rrm2 | -0.053 |
| elavl3 | 0.178 | cx43.4 | -0.053 |
| scg2b | 0.178 | fen1 | -0.053 |
| tmsb | 0.177 | CABZ01005379.1 | -0.052 |
| ppp1r14ba | 0.177 | tspan7 | -0.052 |
| stmn4l | 0.176 | rps20 | -0.052 |
| stxbp1a | 0.176 | mibp | -0.052 |
| sncb | 0.175 | rpa3 | -0.051 |