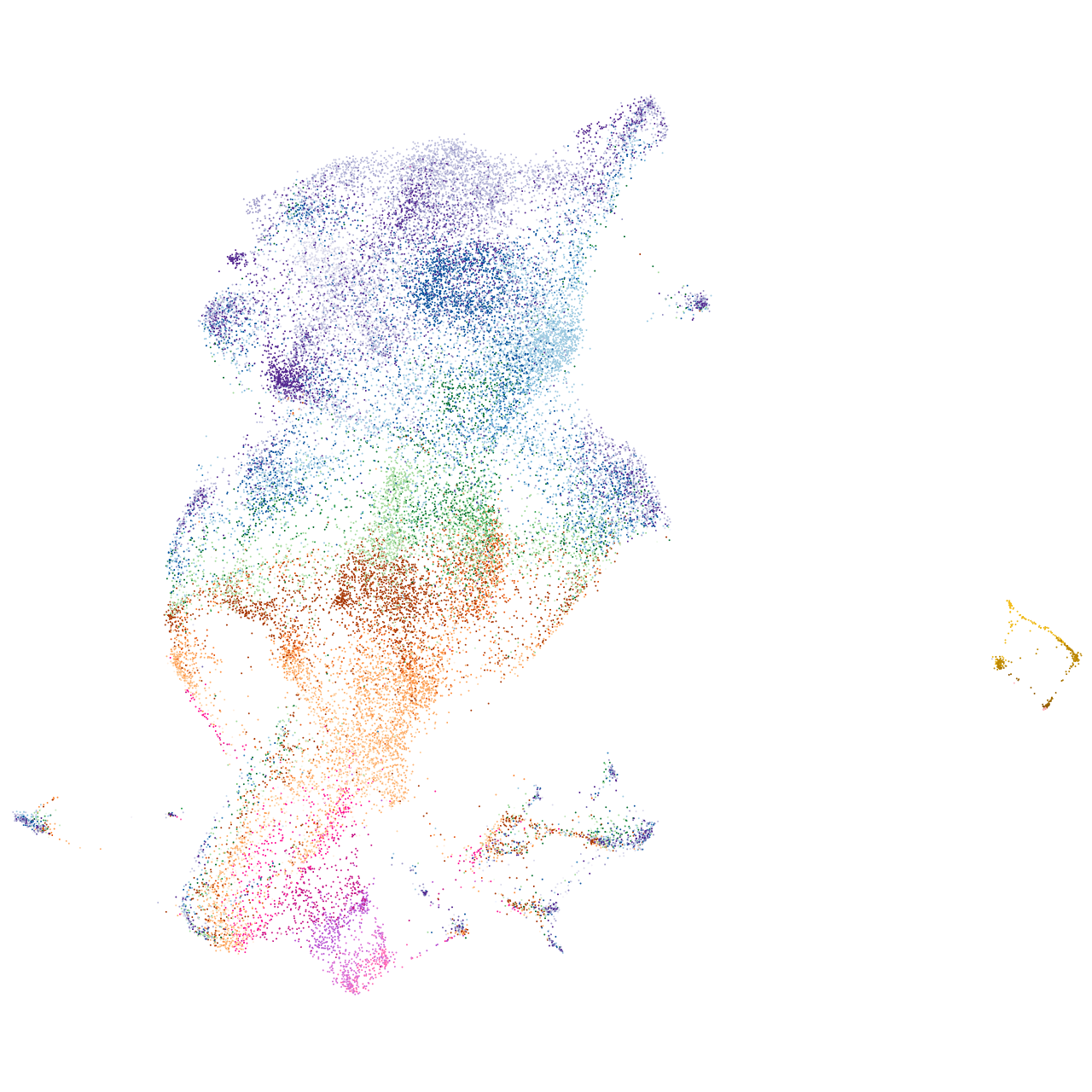Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle 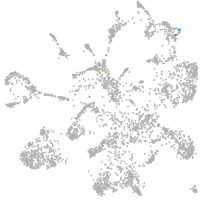 |
mesenchyme 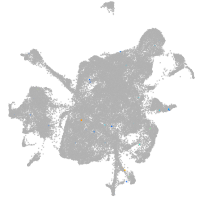 |
hematopoietic / vasculature 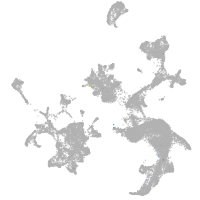 |
pronephros 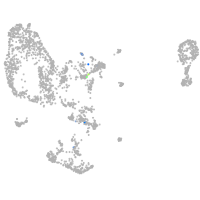 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| evplb | 0.154 | rpl39 | -0.113 |
| cast | 0.140 | romo1 | -0.113 |
| lye | 0.137 | rpl22 | -0.102 |
| mt-cyb | 0.137 | rpl35 | -0.100 |
| si:cabz01007794.1 | 0.132 | ahcy | -0.099 |
| cdh1 | 0.131 | eef1b2 | -0.099 |
| ppl | 0.131 | rpl19 | -0.098 |
| actn4 | 0.127 | rpl7 | -0.098 |
| CR383676.1 | 0.126 | rsl24d1 | -0.096 |
| evpla | 0.124 | rps12 | -0.096 |
| epcam | 0.124 | krtt1c19e | -0.094 |
| dnase1l4.1 | 0.122 | faua | -0.092 |
| CABZ01072614.1 | 0.122 | eif4ebp3 | -0.090 |
| mt-co2 | 0.120 | btf3 | -0.090 |
| cldn7b | 0.120 | rpl10 | -0.089 |
| f11r.1 | 0.117 | rps2 | -0.087 |
| slc25a55a | 0.117 | atp5mc3b | -0.086 |
| mt-nd1 | 0.116 | pno1 | -0.085 |
| mt-atp6 | 0.115 | rpl21 | -0.085 |
| gmds | 0.114 | rps21 | -0.085 |
| zgc:194839 | 0.113 | rpl13a | -0.084 |
| capn9 | 0.110 | rps5 | -0.084 |
| mt-nd4 | 0.109 | rps23 | -0.084 |
| si:ch73-347e22.8 | 0.108 | rps26l | -0.084 |
| hmga1a | 0.108 | rps15a | -0.083 |
| COX3 | 0.106 | glo1 | -0.082 |
| si:dkey-87o1.2 | 0.105 | rplp0 | -0.081 |
| cldne | 0.105 | eevs | -0.081 |
| ildr1a | 0.105 | rpl36a | -0.081 |
| btr12 | 0.105 | rps7 | -0.081 |
| ldlrb | 0.105 | rps29 | -0.081 |
| NC-002333.17 | 0.104 | rpl10a | -0.081 |
| glulb | 0.103 | rps20 | -0.080 |
| tjp3 | 0.103 | gtpbp4 | -0.080 |
| mt-nd2 | 0.103 | rpl8 | -0.080 |