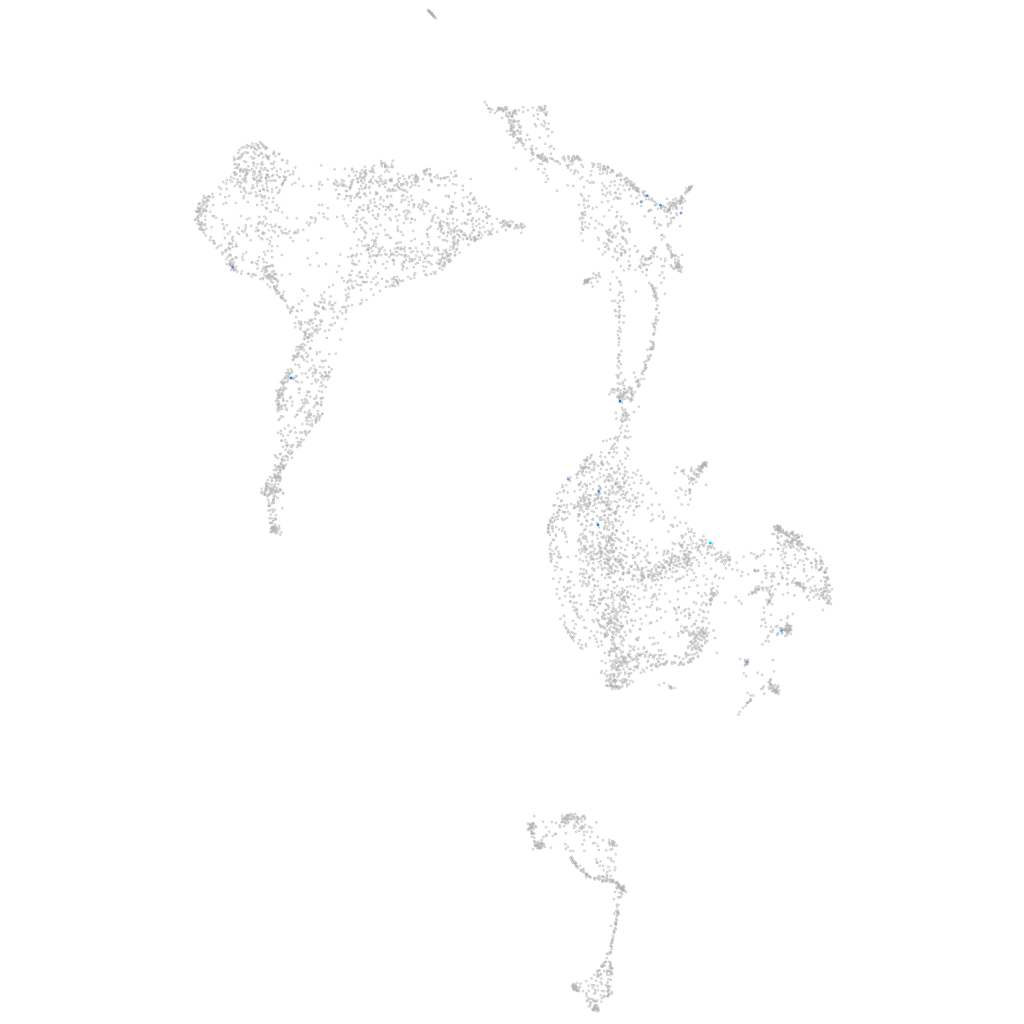
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| drd1a | 0.422 | srsf4 | -0.026 |
| stmnd1 | 0.351 | serbp1b | -0.025 |
| lama2 | 0.348 | phf5a | -0.024 |
| LOC795766 | 0.343 | chrac1 | -0.024 |
| XLOC-001430 | 0.332 | fbl | -0.024 |
| si:ch73-103l1.2 | 0.328 | noto | -0.023 |
| LOC103910643 | 0.313 | prpf3 | -0.023 |
| AL929304.1 | 0.268 | nap1l4a | -0.022 |
| si:ch211-241e1.3 | 0.255 | rbm17 | -0.022 |
| ndufa4l2a | 0.251 | eif3ba | -0.021 |
| calcrla | 0.244 | cirbpa | -0.021 |
| cfap300 | 0.224 | dhx9 | -0.021 |
| hmha1a | 0.219 | si:dkey-177p2.6 | -0.021 |
| kcne4 | 0.214 | sap18 | -0.021 |
| BX649384.1 | 0.212 | rsl1d1 | -0.020 |
| CABZ01084347.1 | 0.212 | yy1a | -0.020 |
| si:dkey-9i23.6 | 0.203 | mfap1 | -0.020 |
| cables2b | 0.196 | hic1l | -0.020 |
| rasl12 | 0.192 | LEPROTL1 | -0.020 |
| ucn3l | 0.186 | rbbp6 | -0.020 |
| entpd2a.1 | 0.183 | mbd3a | -0.020 |
| tm4sf18 | 0.181 | prpf4ba | -0.020 |
| exoc3l1 | 0.180 | mrps34 | -0.020 |
| lmod3 | 0.179 | twistnb | -0.020 |
| CR383662.3 | 0.169 | NC-002333.4 | -0.020 |
| LOC100005685 | 0.167 | nop53 | -0.020 |
| LOC110439551 | 0.167 | npm1a | -0.020 |
| CR735102.1 | 0.165 | cops8 | -0.020 |
| il10 | 0.158 | si:dkey-1j5.4 | -0.020 |
| CT027756.1 | 0.157 | rpl7l1 | -0.019 |
| BX957274.1 | 0.156 | timm10 | -0.019 |
| gba3 | 0.156 | mvb12ba | -0.019 |
| pear1 | 0.156 | selenot1b | -0.019 |
| ednraa | 0.156 | fbxl6 | -0.019 |
| bgnb | 0.156 | admp | -0.019 |