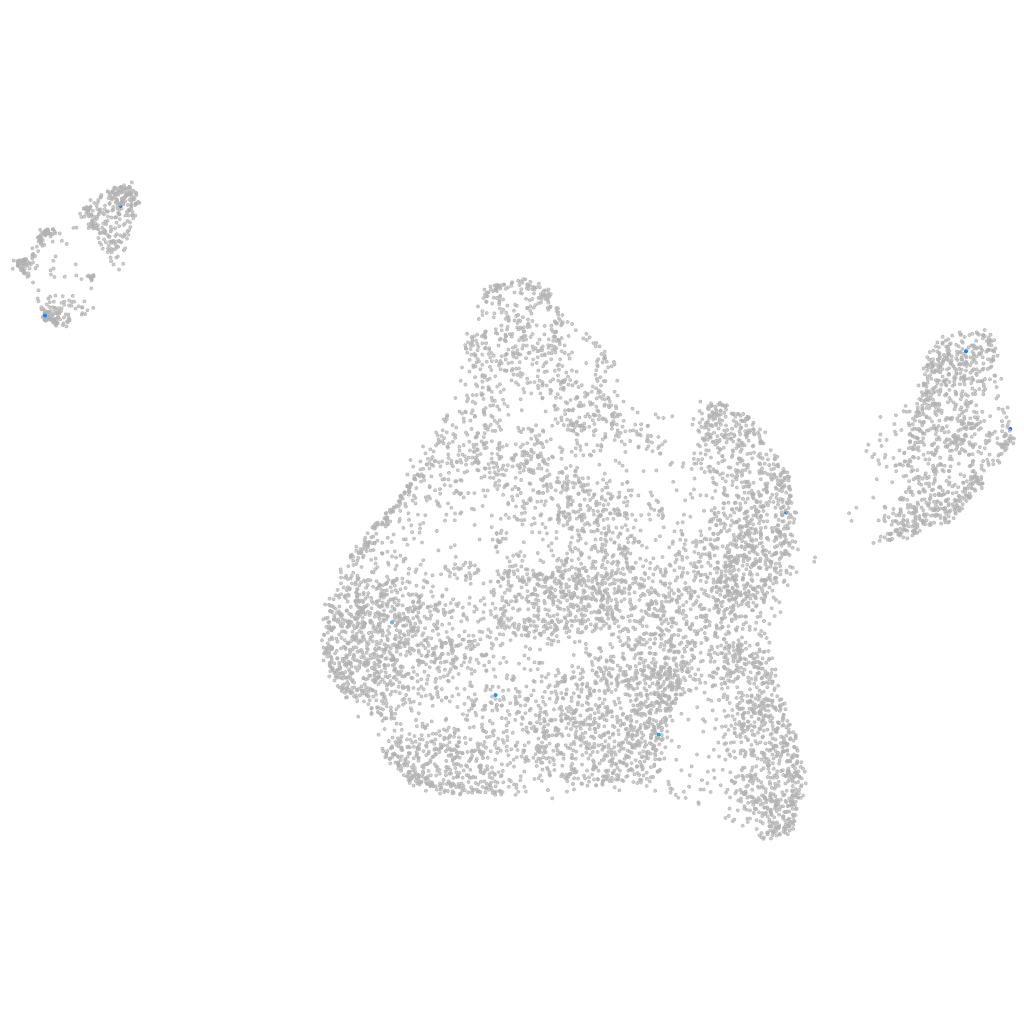
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slc6a17 | 0.224 | ebna1bp2 | -0.026 |
| LOC100536607 | 0.216 | calm2b | -0.026 |
| slit1b | 0.215 | srsf3b | -0.024 |
| zmp:0000000937 | 0.214 | itm2ba | -0.023 |
| LOC100001589 | 0.195 | sf3b1 | -0.023 |
| XLOC-023279 | 0.193 | ptmab | -0.022 |
| XLOC-005677 | 0.191 | tax1bp3 | -0.021 |
| CABZ01084347.1 | 0.168 | rplp1 | -0.020 |
| kcnv2a | 0.164 | alcamb | -0.020 |
| zgc:158328 | 0.164 | rps3 | -0.020 |
| swap70a | 0.163 | gpc4 | -0.019 |
| BX681417.2 | 0.162 | ppil3 | -0.019 |
| agla | 0.138 | add3b | -0.019 |
| st6galnac5a | 0.136 | rpl35 | -0.019 |
| AL929048.1 | 0.134 | srp9 | -0.019 |
| CABZ01046087.1 | 0.131 | gnl2 | -0.019 |
| RF00412 | 0.130 | tgif1 | -0.019 |
| map4l | 0.128 | znf1041 | -0.019 |
| c1qtnf12 | 0.126 | crip1 | -0.019 |
| BX510922.2 | 0.125 | grpel2 | -0.019 |
| atp1b2b | 0.125 | oip5-as1 | -0.019 |
| fhdc4 | 0.121 | atf4a | -0.018 |
| si:ch211-194h1.2 | 0.118 | cops9 | -0.018 |
| gdnfa | 0.116 | si:dkey-286j15.1 | -0.018 |
| muc5e | 0.112 | srsf1b | -0.018 |
| prr33 | 0.111 | rps2 | -0.018 |
| LOC100334739 | 0.110 | u2af2a | -0.018 |
| tnc | 0.110 | smc1al | -0.018 |
| TULP1 | 0.108 | map1lc3b | -0.018 |
| zgc:86896 | 0.107 | hes6 | -0.018 |
| taar20i | 0.107 | dbi | -0.018 |
| ms4a17a.2 | 0.107 | myl12.1 | -0.018 |
| CABZ01071903.1 | 0.105 | wdr46 | -0.018 |
| ifih1 | 0.104 | mgaa | -0.018 |
| BX649556.1 | 0.103 | mrto4 | -0.018 |