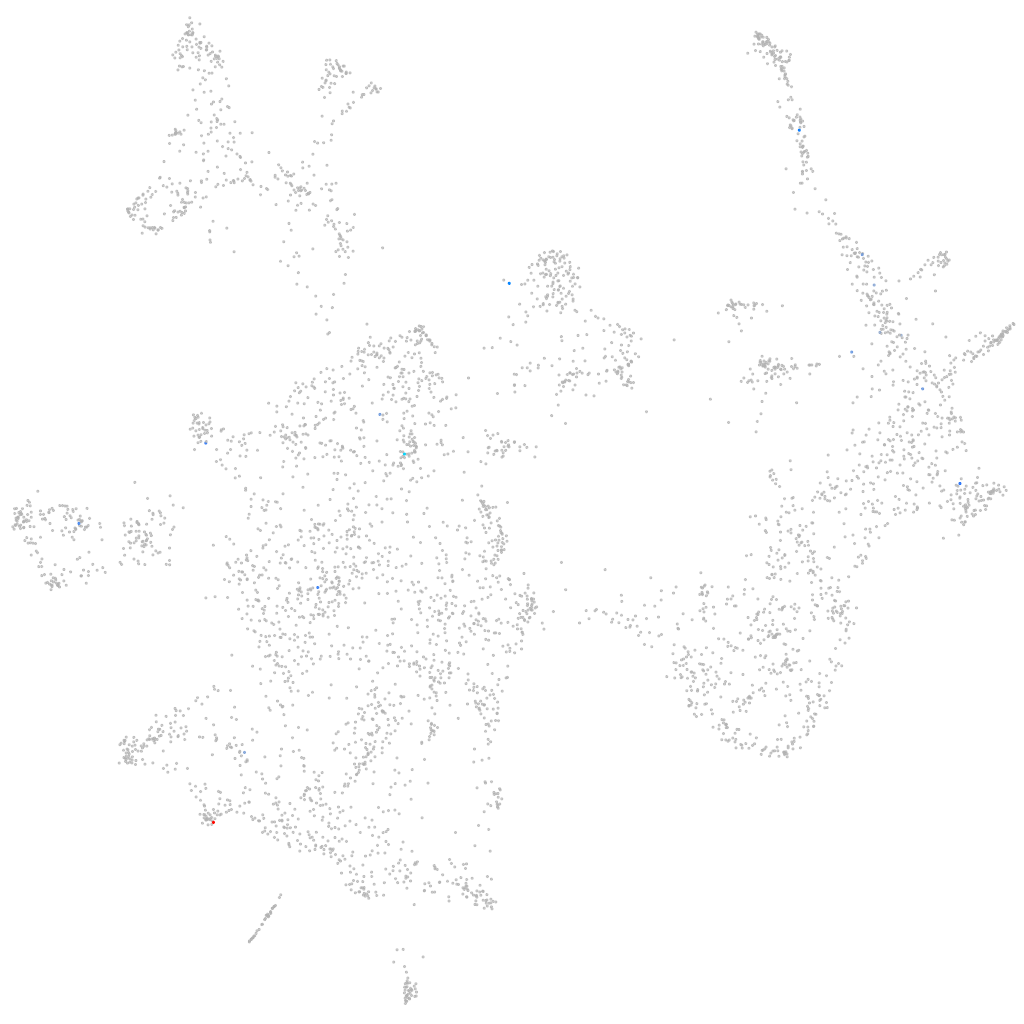
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC101886946.1 | 0.774 | hspa8 | -0.074 |
| CR762407.1 | 0.660 | rps15 | -0.070 |
| CABZ01067979.1 | 0.561 | rpl23 | -0.070 |
| irak3 | 0.443 | rps2 | -0.067 |
| LOC110437714 | 0.439 | rps7 | -0.063 |
| zgc:103438 | 0.434 | rpl35 | -0.063 |
| FP102018.1 | 0.434 | rps23 | -0.063 |
| mmp11b | 0.405 | rps13 | -0.059 |
| LOC110438251 | 0.379 | rps9 | -0.059 |
| cdc42ep1a | 0.369 | rps28 | -0.059 |
| ppfibp2b | 0.361 | rpl10 | -0.059 |
| zgc:101540 | 0.350 | rps21 | -0.059 |
| ric1 | 0.321 | faua | -0.059 |
| lratb.2 | 0.302 | rpl18 | -0.056 |
| fam129ba | 0.302 | rps4x | -0.056 |
| ciz1b | 0.285 | rpl17 | -0.055 |
| zbtb4 | 0.271 | rps26l | -0.055 |
| agpat2 | 0.267 | rpl22 | -0.054 |
| klhl22 | 0.263 | rpl10a | -0.053 |
| fgf8b | 0.260 | eef1b2 | -0.052 |
| BX842699.1 | 0.258 | rps12 | -0.051 |
| VIT | 0.250 | rps27.2 | -0.049 |
| atp7a | 0.249 | rack1 | -0.045 |
| stx10 | 0.238 | naca | -0.044 |
| elk3 | 0.229 | ybx1 | -0.043 |
| nrg1 | 0.229 | rps26 | -0.041 |
| ino80da | 0.228 | rpl5a | -0.041 |
| thbs1a | 0.226 | mt-atp6 | -0.040 |
| vwa8 | 0.222 | h3f3d | -0.040 |
| mlycd | 0.219 | rpl9 | -0.040 |
| jarid2a | 0.217 | ppiab | -0.039 |
| ago1 | 0.217 | cox4i1 | -0.038 |
| CR457445.1 | 0.212 | ndufa4l | -0.038 |
| LOC103911969 | 0.208 | atp5mc3b | -0.037 |
| ada2a | 0.208 | eef2b | -0.037 |