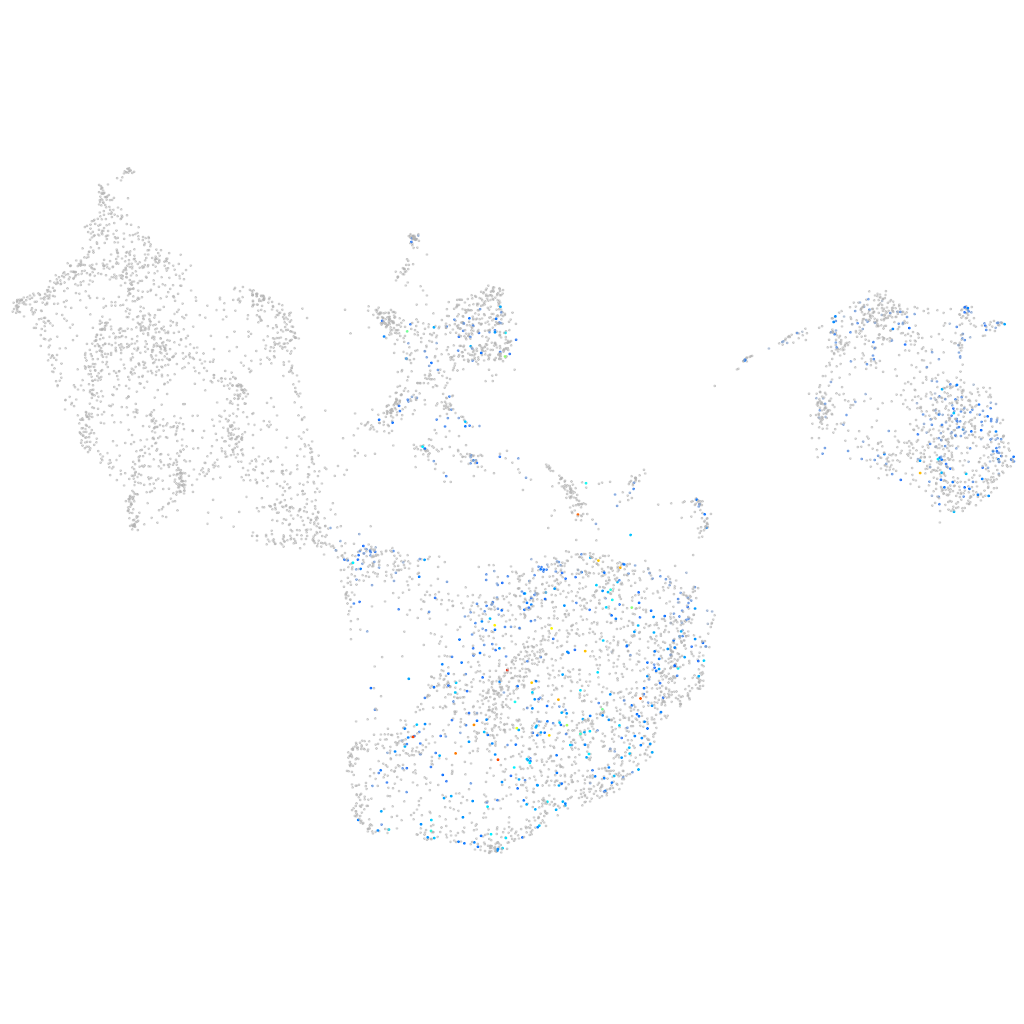
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cldni | 0.223 | c1qtnf5 | -0.158 |
| myh9a | 0.223 | bhmt | -0.148 |
| si:dkey-102c8.3 | 0.221 | fbln1 | -0.145 |
| epcam | 0.214 | hgd | -0.142 |
| pfn1 | 0.213 | cd82a | -0.141 |
| cldn1 | 0.212 | ttc36 | -0.134 |
| col18a1a | 0.212 | tmem119b | -0.132 |
| zgc:92380 | 0.210 | rplp1 | -0.129 |
| tmsb1 | 0.209 | cfl1 | -0.129 |
| cfl1l | 0.208 | prrx1a | -0.129 |
| tagln2 | 0.208 | pcbd1 | -0.129 |
| spaca4l | 0.205 | twist1a | -0.127 |
| apoeb | 0.204 | si:dkey-151g10.6 | -0.126 |
| egfl6 | 0.204 | rplp2l | -0.126 |
| rac2 | 0.203 | mfap2 | -0.125 |
| bcam | 0.202 | qdpra | -0.125 |
| cotl1 | 0.201 | ptx3a | -0.125 |
| thy1 | 0.200 | rpl39 | -0.124 |
| fmodb | 0.198 | hpdb | -0.124 |
| and3 | 0.198 | prrx1b | -0.122 |
| itgb1b.1 | 0.195 | rpl37 | -0.121 |
| spint1a | 0.194 | crabp2a | -0.121 |
| arhgdig | 0.193 | nme2b.1 | -0.120 |
| plecb | 0.193 | gpc1a | -0.119 |
| tp63 | 0.192 | msna | -0.116 |
| tmsb4x | 0.190 | rpl12 | -0.116 |
| anxa1a | 0.189 | reck | -0.116 |
| s100a10b | 0.188 | rpl36a | -0.116 |
| ptgs2a | 0.188 | rps15a | -0.115 |
| si:rp71-77l1.1 | 0.187 | fah | -0.114 |
| lama5 | 0.186 | rps21 | -0.114 |
| zgc:101810 | 0.186 | twist3 | -0.113 |
| cap1 | 0.185 | calr | -0.112 |
| lgals3b | 0.182 | mdh1aa | -0.110 |
| si:dkey-248g15.3 | 0.182 | ntd5 | -0.110 |