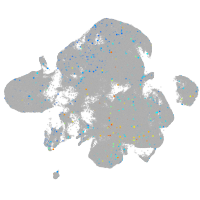
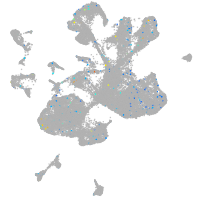
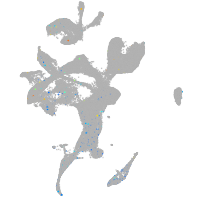
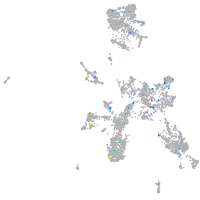
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CT573186.2 | 0.039 | hmga1a | -0.024 |
| calm1b | 0.039 | cirbpb | -0.022 |
| rab6bb | 0.037 | hnrnpub | -0.021 |
| pimr168 | 0.037 | cx43.4 | -0.020 |
| uchl1 | 0.036 | si:ch211-222l21.1 | -0.019 |
| gapdhs | 0.035 | hmgb2a | -0.018 |
| adgre14 | 0.033 | khdrbs1a | -0.018 |
| ywhag2 | 0.033 | hmgb2b | -0.017 |
| atp6v1e1b | 0.033 | FO082781.1 | -0.016 |
| zgc:65894 | 0.032 | eef1a1l1 | -0.016 |
| eno1a | 0.032 | cirbpa | -0.016 |
| sncgb | 0.032 | rbm4.3 | -0.016 |
| atp6v1g1 | 0.032 | NC-002333.4 | -0.016 |
| SMIM18 | 0.031 | hnrnpa1b | -0.016 |
| tmsb2 | 0.031 | stm | -0.015 |
| eno2 | 0.031 | sept12 | -0.015 |
| ywhag1 | 0.031 | pfkfb4b | -0.015 |
| syn2a | 0.030 | ilf3b | -0.015 |
| stxbp1a | 0.030 | pou5f3 | -0.015 |
| CR450824.2 | 0.030 | hspb1 | -0.015 |
| tpi1b | 0.030 | gar1 | -0.014 |
| b4galnt2.1 | 0.030 | hmgn2 | -0.014 |
| calm3a | 0.030 | si:ch211-152c2.3 | -0.014 |
| stmn2a | 0.029 | apoeb | -0.014 |
| cdk5r2a | 0.029 | rrp15 | -0.014 |
| pkma | 0.029 | matr3l1.1 | -0.014 |
| LOC101882847 | 0.029 | eif4bb | -0.014 |
| drd6b | 0.029 | rps20 | -0.014 |
| calm2a | 0.028 | smarcd1 | -0.014 |
| calm1a | 0.028 | nop58 | -0.014 |
| gabarapl2 | 0.028 | srsf1a | -0.013 |
| XLOC-037389 | 0.028 | crabp2b | -0.013 |
| gng3 | 0.028 | apoc1 | -0.013 |
| sncb | 0.027 | rac1a | -0.013 |
| jpt1a | 0.027 | polr3gla | -0.013 |