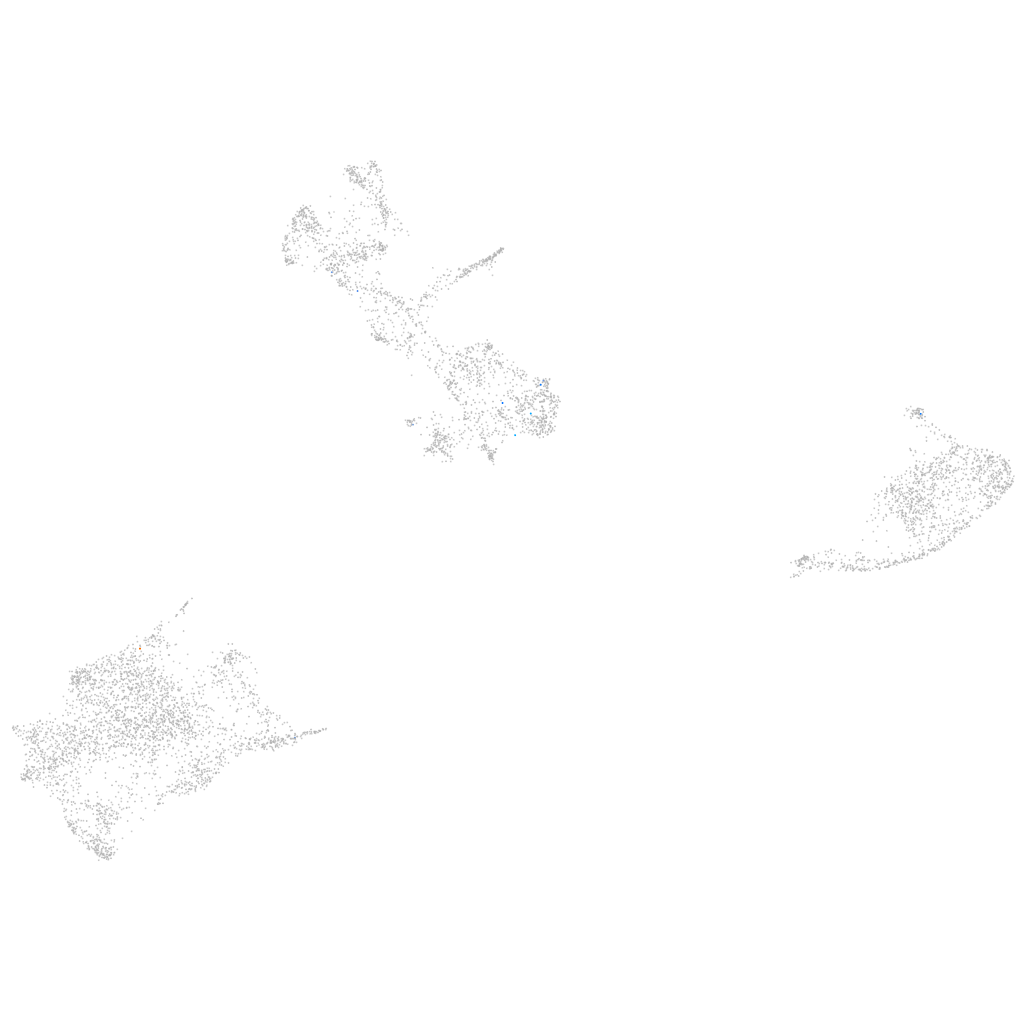
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural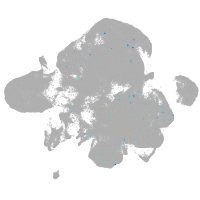 |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line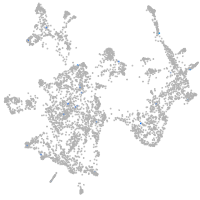 |
epidermis |
periderm |
fin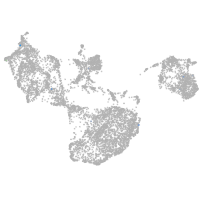 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| crygm2d3 | 0.399 | rps24 | -0.050 |
| def6c | 0.394 | rps27a | -0.045 |
| mocs2 | 0.345 | rpl11 | -0.042 |
| si:dkey-50i6.6 | 0.296 | rpl8 | -0.041 |
| adra2db | 0.256 | rps10 | -0.041 |
| rbks | 0.250 | rps29 | -0.039 |
| trabd2a | 0.242 | rps7 | -0.039 |
| apex2 | 0.239 | rpl18 | -0.036 |
| nxph3 | 0.222 | rpl21 | -0.036 |
| CR589947.3 | 0.208 | rpl14 | -0.036 |
| anapc1 | 0.205 | rpl27 | -0.035 |
| FQ311920.1 | 0.191 | rps3a | -0.035 |
| si:dkey-13p1.4 | 0.182 | rpl31 | -0.035 |
| si:ch1073-429i10.3 | 0.180 | rpl18a | -0.032 |
| si:dkey-117i10.1 | 0.180 | rps5 | -0.031 |
| trim35-19 | 0.176 | rps4x | -0.030 |
| tmem56a | 0.175 | rps11 | -0.030 |
| atf7ip2 | 0.174 | zgc:114188 | -0.030 |
| elf3 | 0.172 | gstp1 | -0.030 |
| zgc:103508 | 0.168 | rpl34 | -0.028 |
| BX601648.1 | 0.167 | rpl22 | -0.027 |
| dusp5 | 0.163 | rps21 | -0.027 |
| FQ377991.1 | 0.162 | actb2 | -0.026 |
| tbrg1 | 0.159 | rpl23a | -0.025 |
| trhrb | 0.157 | rack1 | -0.024 |
| si:dkey-280e21.3 | 0.157 | pcbd1 | -0.024 |
| si:ch73-306e8.2 | 0.152 | rplp2 | -0.023 |
| znf281a | 0.151 | cst14a.2 | -0.022 |
| nr4a1 | 0.149 | pfn1 | -0.022 |
| tmem184ba | 0.148 | qdpra | -0.022 |
| rgs14a | 0.146 | paics | -0.021 |
| creld2 | 0.145 | pts | -0.021 |
| phldb2a | 0.145 | gpr143 | -0.021 |
| LOC110438108 | 0.140 | csrp2 | -0.020 |
| casq1a | 0.140 | mdh1aa | -0.020 |