Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme 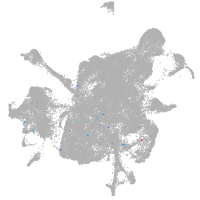 |
hematopoietic / vasculature 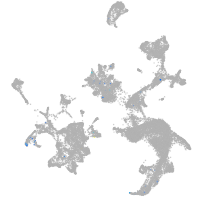 |
pronephros  |
neural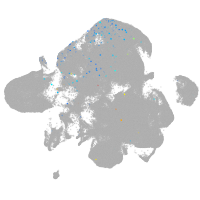 |
spinal cord / glia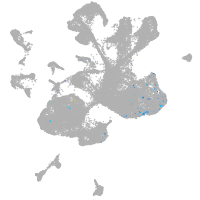 |
eye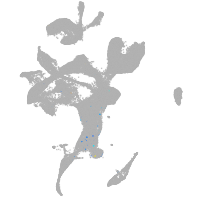 |
taste / olfactory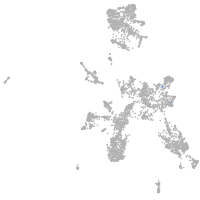 |
otic / lateral line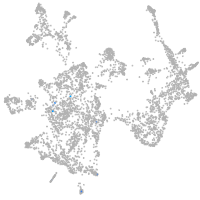 |
epidermis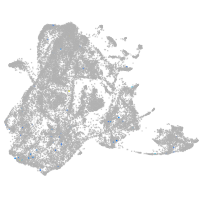 |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gmnn | 0.067 | elavl3 | -0.044 |
| pcna | 0.066 | rtn1a | -0.044 |
| fam177b | 0.064 | ckbb | -0.040 |
| CABZ01075068.1 | 0.063 | stmn1b | -0.039 |
| dut | 0.062 | gpm6ab | -0.035 |
| selenoh | 0.061 | gpm6aa | -0.035 |
| msh6 | 0.059 | tmsb | -0.035 |
| rpa2 | 0.057 | gng3 | -0.033 |
| XLOC-032526 | 0.056 | sncb | -0.032 |
| mcm7 | 0.056 | rtn1b | -0.032 |
| si:dkey-7j22.1 | 0.056 | atp6v0cb | -0.031 |
| nutf2l | 0.056 | hmgb3a | -0.031 |
| fen1 | 0.056 | rnasekb | -0.031 |
| rrm2 | 0.055 | COX3 | -0.030 |
| XLOC-008823 | 0.055 | tuba1c | -0.030 |
| zgc:56493 | 0.053 | fez1 | -0.030 |
| shmt1 | 0.053 | pvalb1 | -0.029 |
| mcm5 | 0.053 | tubb5 | -0.029 |
| gins2 | 0.053 | elavl4 | -0.029 |
| her2 | 0.053 | myt1b | -0.029 |
| unga | 0.052 | stmn2a | -0.028 |
| rpa3 | 0.052 | vamp2 | -0.028 |
| XLOC-003690 | 0.052 | ywhag2 | -0.028 |
| id1 | 0.051 | stx1b | -0.028 |
| rrm1 | 0.051 | hbbe1.3 | -0.028 |
| mcm4 | 0.051 | si:dkeyp-75h12.5 | -0.028 |
| cldn5a | 0.051 | dpysl3 | -0.028 |
| chtf8 | 0.051 | ube2d4 | -0.028 |
| cdc6 | 0.050 | kdm6bb | -0.028 |
| gfap | 0.050 | marcksl1b | -0.028 |
| her9 | 0.050 | hbae3 | -0.028 |
| paics | 0.050 | hmgb1a | -0.027 |
| orc4 | 0.050 | mllt11 | -0.027 |
| LOC100330864 | 0.050 | zgc:65894 | -0.027 |
| mcm2 | 0.049 | gap43 | -0.026 |