Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 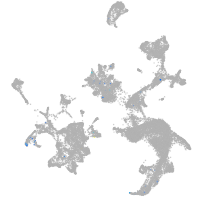 |
pronephros  |
neural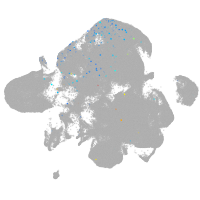 |
spinal cord / glia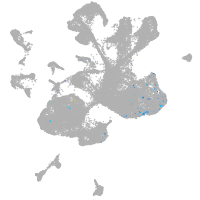 |
eye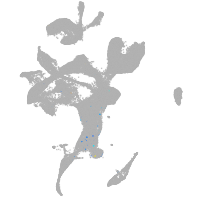 |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pcna | 0.061 | pvalb1 | -0.022 |
| rpa2 | 0.059 | pvalb2 | -0.021 |
| dut | 0.058 | actc1b | -0.020 |
| fen1 | 0.058 | ckbb | -0.017 |
| XLOC-032526 | 0.057 | tmsb | -0.017 |
| mcm7 | 0.056 | col1a2 | -0.016 |
| gmnn | 0.056 | eef1da | -0.016 |
| mcm5 | 0.055 | col1a1a | -0.015 |
| CABZ01075068.1 | 0.054 | elavl3 | -0.015 |
| mcm4 | 0.053 | mylpfa | -0.015 |
| orc4 | 0.052 | myt1b | -0.015 |
| selenoh | 0.052 | col1a1b | -0.014 |
| nutf2l | 0.051 | cotl1 | -0.014 |
| gins2 | 0.050 | krt8 | -0.014 |
| rpa3 | 0.050 | rtn1a | -0.014 |
| unga | 0.050 | scrt2 | -0.014 |
| rrm2 | 0.050 | si:dkey-28b4.7 | -0.014 |
| cad | 0.049 | bhmt | -0.014 |
| chtf8 | 0.049 | col2a1b | -0.013 |
| mcm2 | 0.049 | dcn | -0.013 |
| mcm6 | 0.049 | icn | -0.013 |
| rnaseh2a | 0.049 | myt1a | -0.013 |
| cdca7b | 0.048 | ndrg2 | -0.013 |
| cnbpa | 0.048 | stmn1b | -0.013 |
| her2 | 0.048 | zgc:162730 | -0.013 |
| mcm3 | 0.048 | col5a1 | -0.013 |
| ranbp1 | 0.048 | ebf3a | -0.013 |
| rrm1 | 0.048 | CU929237.1 | -0.012 |
| shmt1 | 0.048 | gpm6ab | -0.012 |
| nasp | 0.047 | hbbe1.3 | -0.012 |
| zgc:110540 | 0.047 | ip6k2a | -0.012 |
| CU929070.1 | 0.046 | kdm6bb | -0.012 |
| dhfr | 0.046 | krt18b | -0.012 |
| hells | 0.046 | krt91 | -0.012 |
| LOC100330864 | 0.046 | krtt1c19e | -0.012 |




