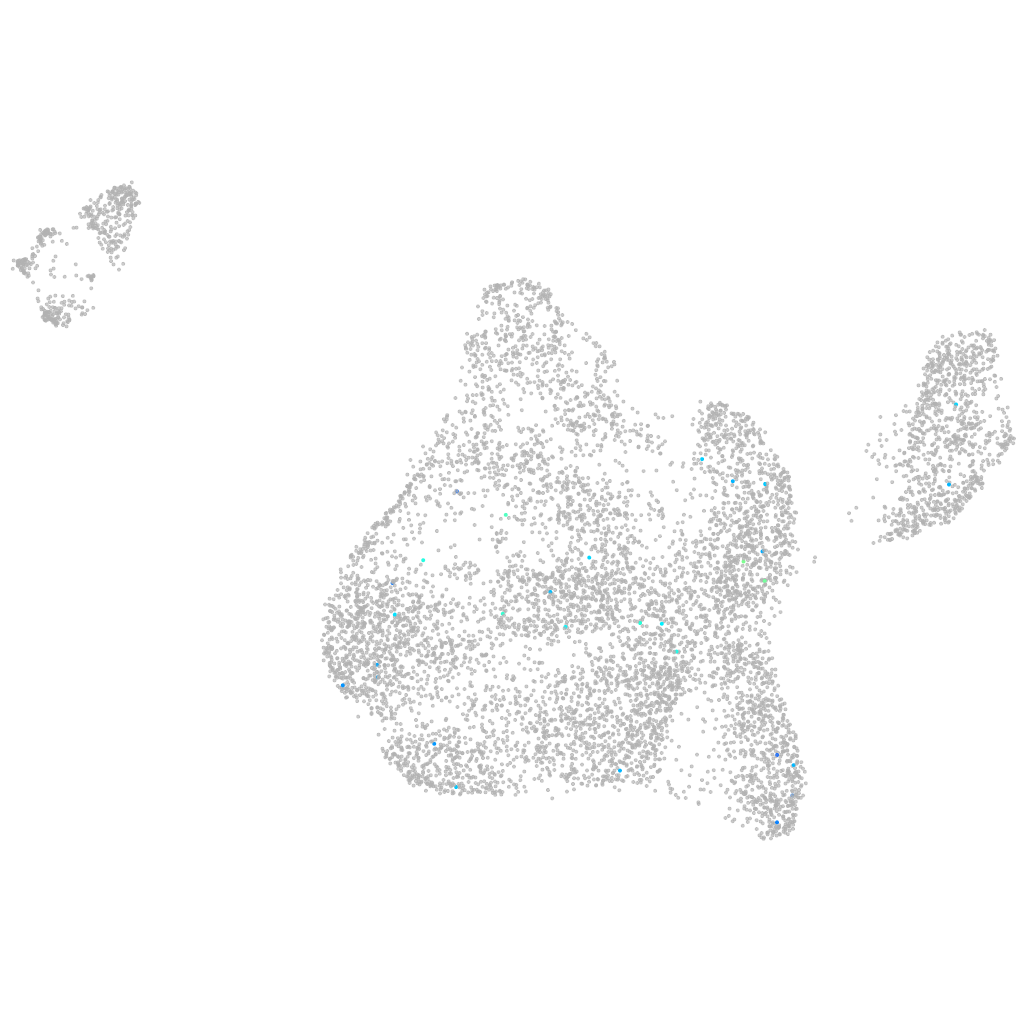
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tgm8 | 0.242 | sf3a2 | -0.033 |
| XLOC-027877 | 0.231 | dmap1 | -0.031 |
| LOC558761 | 0.182 | hmg20b | -0.030 |
| LOC108182924 | 0.182 | arpp19b | -0.028 |
| lrrfip1b | 0.180 | hdgfl2 | -0.027 |
| tmem204 | 0.178 | ctsc | -0.027 |
| BX530064.1 | 0.175 | eif4e1b | -0.026 |
| inab | 0.175 | zgc:101744 | -0.026 |
| CT025669.1 | 0.170 | astn1 | -0.026 |
| LOC100536487 | 0.169 | ankrd12 | -0.026 |
| si:dkey-205h23.2 | 0.165 | atp1b3b | -0.026 |
| diras1a | 0.160 | atp6v0ca | -0.026 |
| si:ch211-69e13.6 | 0.160 | tfap4 | -0.025 |
| neu3.3 | 0.156 | akirin1 | -0.025 |
| LOC101882272 | 0.154 | tmem258 | -0.025 |
| tmem200a | 0.146 | hectd1 | -0.025 |
| cpeb1a | 0.140 | arfgap3 | -0.025 |
| RF01233 | 0.140 | gcat | -0.024 |
| CT573408.1 | 0.139 | dolpp1 | -0.024 |
| mhc1uma | 0.138 | smim12 | -0.024 |
| ky | 0.136 | ppwd1 | -0.024 |
| CU207241.1 | 0.132 | chp1 | -0.024 |
| si:dkeyp-86c4.1 | 0.127 | strn | -0.024 |
| XLOC-020173 | 0.123 | ccnb2 | -0.024 |
| slc6a17 | 0.123 | commd7 | -0.023 |
| gas2l2 | 0.121 | stk40 | -0.023 |
| adra2c | 0.120 | rpl5b | -0.023 |
| LOC110438594 | 0.115 | prkacbb | -0.023 |
| znf362b | 0.113 | gtf3ab | -0.023 |
| spic | 0.112 | cdk8 | -0.023 |
| enam | 0.111 | elovl1b | -0.022 |
| pimr93 | 0.110 | timm29 | -0.022 |
| galnt18b | 0.106 | lyar | -0.022 |
| XLOC-019772 | 0.105 | atp5meb | -0.022 |
| ak5 | 0.104 | gskip | -0.022 |