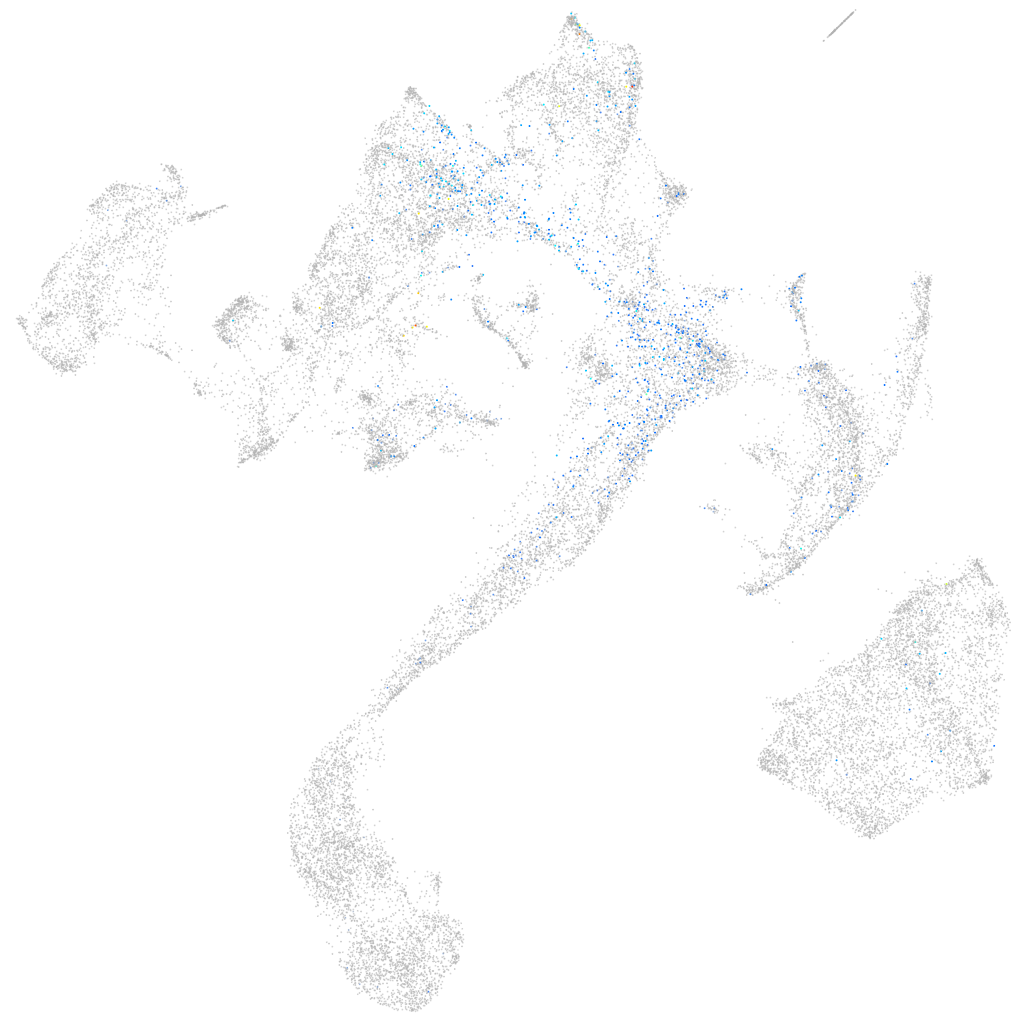
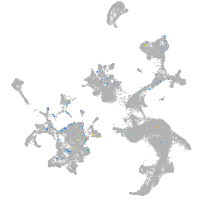
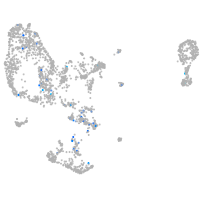
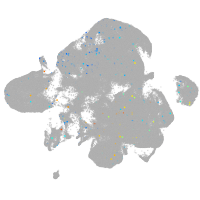
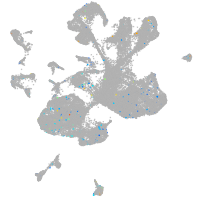
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-43f4.1 | 0.183 | gapdh | -0.087 |
| mdka | 0.157 | ckma | -0.085 |
| lrrc17 | 0.154 | tnnc2 | -0.085 |
| vwde | 0.151 | ak1 | -0.084 |
| pax3a | 0.150 | aldoab | -0.084 |
| emp2 | 0.148 | cycsb | -0.083 |
| angptl7 | 0.142 | atp2a1 | -0.083 |
| postnb | 0.141 | tmem38a | -0.083 |
| tcf15 | 0.140 | neb | -0.082 |
| twist1a | 0.139 | ckmb | -0.081 |
| ssr3 | 0.138 | desma | -0.081 |
| col5a2a | 0.135 | actc1b | -0.080 |
| mfap2 | 0.133 | acta1b | -0.078 |
| dmrt2a | 0.129 | ldb3b | -0.077 |
| uncx4.1 | 0.127 | cox7c | -0.077 |
| XLOC-042229 | 0.120 | acta1a | -0.077 |
| itga10 | 0.118 | actn3a | -0.076 |
| meox1 | 0.115 | mybphb | -0.076 |
| ahnak | 0.115 | ldb3a | -0.076 |
| glula | 0.115 | actn3b | -0.075 |
| thbs4b | 0.114 | si:ch73-367p23.2 | -0.075 |
| nectin1b | 0.114 | tpma | -0.075 |
| itga9 | 0.113 | si:ch211-266g18.10 | -0.074 |
| ssr2 | 0.111 | cav3 | -0.074 |
| pax7a | 0.111 | smyd1a | -0.073 |
| wnt16 | 0.109 | srl | -0.073 |
| ssr4 | 0.109 | myom1a | -0.073 |
| nr2f5 | 0.109 | casq2 | -0.073 |
| col5a1 | 0.109 | CABZ01078594.1 | -0.073 |
| ppib | 0.108 | hhatla | -0.072 |
| aldh1a2 | 0.108 | mylpfa | -0.072 |
| pdia3 | 0.108 | pgam2 | -0.072 |
| her6 | 0.108 | tmod4 | -0.071 |
| kirrel3l | 0.107 | trdn | -0.071 |
| notch2 | 0.107 | stac3 | -0.071 |