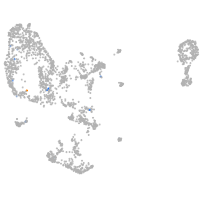
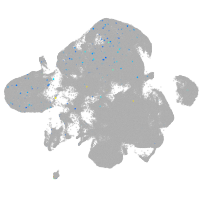
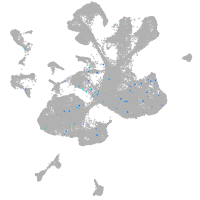
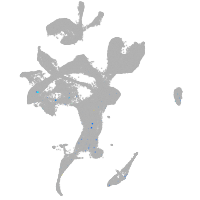
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:136767 | 0.070 | stmn1b | -0.038 |
| FP003590.1 | 0.052 | rtn1a | -0.038 |
| si:ch211-256e16.4 | 0.046 | gng3 | -0.034 |
| AL929586.1 | 0.046 | ckbb | -0.034 |
| chrna4a | 0.045 | elavl3 | -0.033 |
| BX279525.1 | 0.045 | gpm6ab | -0.032 |
| id1 | 0.044 | sncb | -0.031 |
| nfatc2a | 0.043 | atp6v0cb | -0.030 |
| trim35-24 | 0.043 | rnasekb | -0.030 |
| si:dkey-10c21.1 | 0.042 | rtn1b | -0.030 |
| hmgb2a | 0.041 | elavl4 | -0.029 |
| ush1gb | 0.041 | ptmaa | -0.029 |
| ccnd1 | 0.041 | tuba1c | -0.028 |
| pkd1a | 0.040 | atp6v1g1 | -0.028 |
| sdc4 | 0.040 | ywhag2 | -0.028 |
| pcna | 0.039 | mllt11 | -0.028 |
| her2 | 0.039 | stmn2a | -0.028 |
| msna | 0.039 | gpm6aa | -0.028 |
| XLOC-003690 | 0.038 | zgc:65894 | -0.028 |
| stmn1a | 0.038 | snap25a | -0.028 |
| chaf1a | 0.038 | si:dkeyp-75h12.5 | -0.027 |
| XLOC-011415 | 0.038 | calm1a | -0.027 |
| XLOC-023481 | 0.037 | gap43 | -0.027 |
| zgc:110216 | 0.036 | zc4h2 | -0.027 |
| lig1 | 0.036 | vamp2 | -0.027 |
| rrm1 | 0.036 | gng2 | -0.027 |
| pimr47 | 0.036 | ccni | -0.027 |
| ranbp1 | 0.036 | tmsb | -0.027 |
| lbr | 0.035 | atp6v1e1b | -0.027 |
| rrm2 | 0.035 | stxbp1a | -0.026 |
| cldn5a | 0.035 | stx1b | -0.026 |
| mki67 | 0.034 | fez1 | -0.026 |
| akap12b | 0.034 | h2afx1 | -0.026 |
| dut | 0.034 | tmsb2 | -0.026 |
| anp32b | 0.034 | gapdhs | -0.026 |