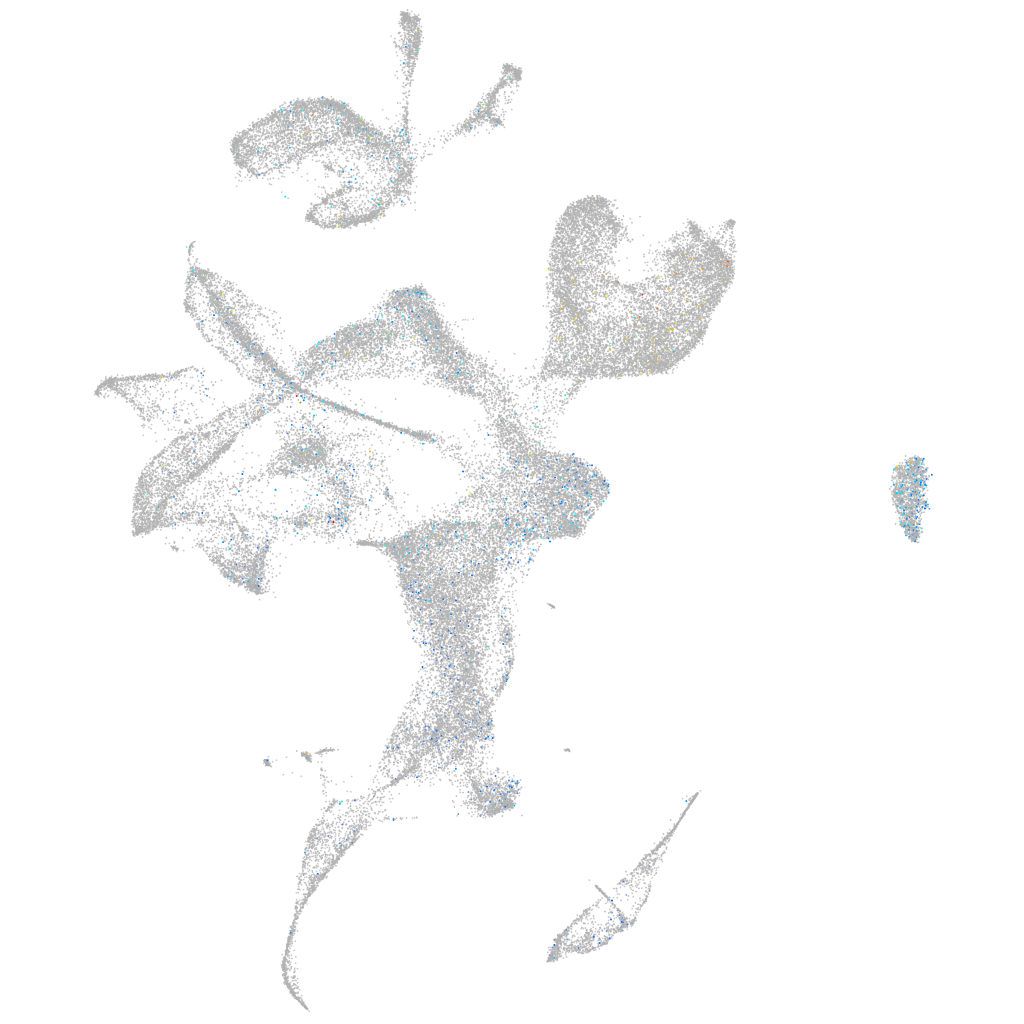
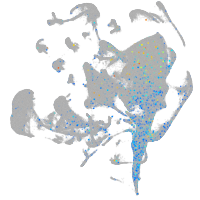
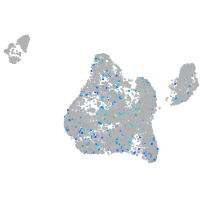
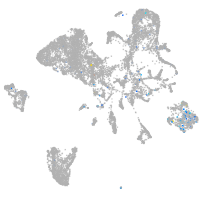
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| shisa2a | 0.064 | gapdhs | -0.030 |
| COX7A2 | 0.062 | pvalb2 | -0.028 |
| hspb1 | 0.060 | mt-atp6 | -0.028 |
| tpx2 | 0.058 | aldocb | -0.027 |
| apoeb | 0.056 | tpi1b | -0.027 |
| ube2c | 0.056 | actc1b | -0.027 |
| nusap1 | 0.056 | apoa2 | -0.026 |
| mad2l1 | 0.056 | si:dkey-16p21.8 | -0.026 |
| mki67 | 0.055 | pvalb1 | -0.026 |
| cdca8 | 0.055 | eno1a | -0.025 |
| stm | 0.055 | cplx4a | -0.024 |
| marcksb | 0.054 | snap25b | -0.024 |
| sox19a | 0.054 | si:ch211-195b13.1 | -0.024 |
| tdgf1 | 0.053 | ak1 | -0.024 |
| nr6a1a | 0.053 | tob1a | -0.024 |
| cenpf | 0.053 | pkma | -0.024 |
| top1l | 0.052 | atp6v0cb | -0.023 |
| alcamb | 0.052 | atp5if1b | -0.023 |
| hmga1a | 0.051 | cox7a2a | -0.023 |
| ccnb1 | 0.051 | gnat2 | -0.023 |
| gig2l | 0.050 | gpia | -0.023 |
| smarca4a | 0.050 | pgk1 | -0.023 |
| cyp26a1 | 0.050 | syt5a | -0.023 |
| hnrnpa1b | 0.049 | sncgb | -0.023 |
| kpna2 | 0.049 | sh3gl2a | -0.023 |
| si:ch211-152c2.3 | 0.049 | atp5l | -0.022 |
| nono | 0.049 | gngt2b | -0.022 |
| top2a | 0.048 | rbp4l | -0.022 |
| chrd | 0.048 | mt-nd1 | -0.022 |
| plk1 | 0.048 | gnb3b | -0.022 |
| LOC100538023 | 0.048 | prss59.2 | -0.022 |
| zmp:0000000624 | 0.048 | si:dkey-17e16.15 | -0.021 |
| prc1a | 0.047 | ldhba | -0.021 |
| kif11 | 0.047 | zgc:73359 | -0.021 |
| hesx1 | 0.047 | prph2b | -0.021 |