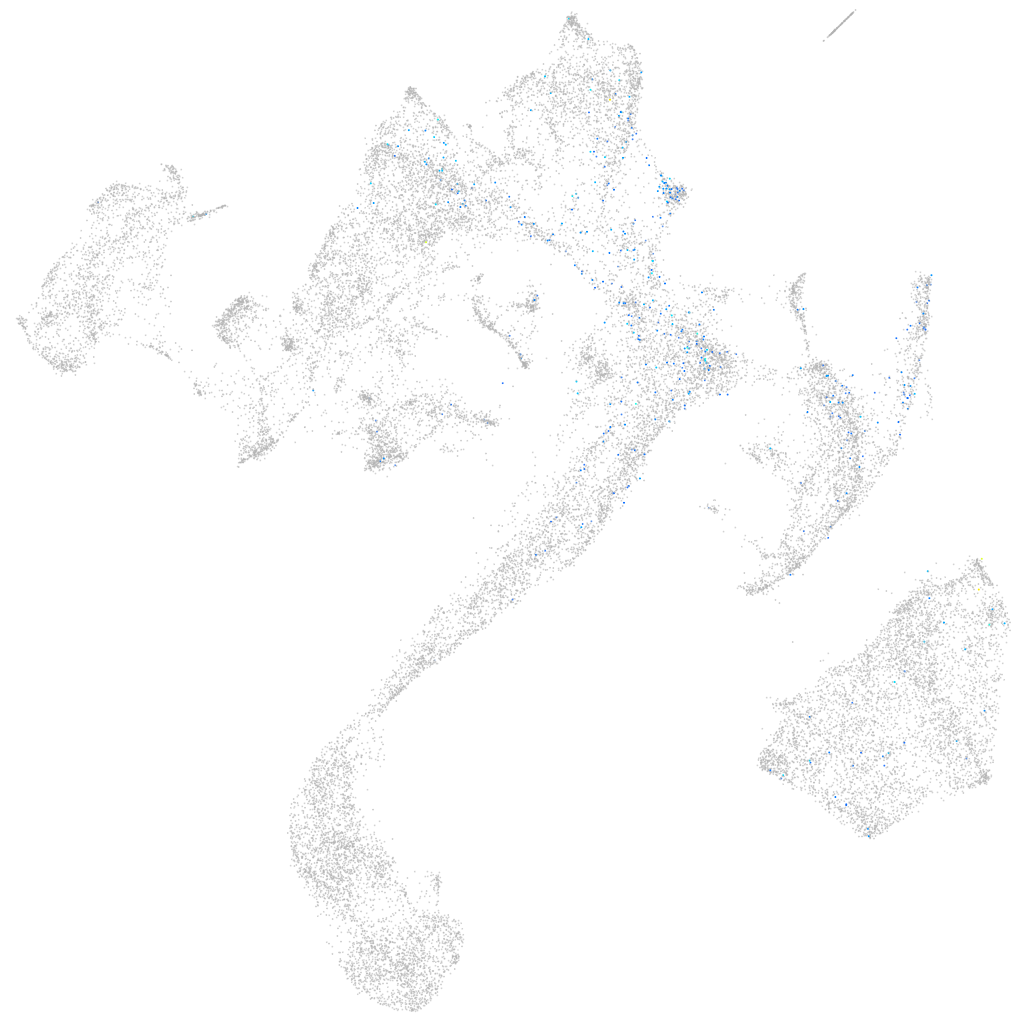
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CU570769.1 | 0.106 | ckma | -0.064 |
| angptl7 | 0.103 | atp2a1 | -0.064 |
| mfap2 | 0.088 | ckmb | -0.063 |
| meox1 | 0.086 | actc1b | -0.059 |
| fstl1a | 0.084 | gapdh | -0.059 |
| mdka | 0.083 | pabpc4 | -0.057 |
| ssr3 | 0.082 | tmem38a | -0.057 |
| sdc4 | 0.080 | ak1 | -0.057 |
| atf3 | 0.080 | ldb3b | -0.056 |
| tpm4a | 0.079 | mylpfa | -0.056 |
| emp2 | 0.079 | tnnc2 | -0.056 |
| CU462871.1 | 0.078 | neb | -0.055 |
| XLOC-042229 | 0.078 | aldoab | -0.055 |
| fn1b | 0.078 | myom1a | -0.054 |
| nkx3-1 | 0.078 | si:ch73-367p23.2 | -0.054 |
| colec12 | 0.078 | mylz3 | -0.053 |
| cpn1 | 0.077 | cox7c | -0.053 |
| lin28a | 0.077 | myl1 | -0.053 |
| tcf15 | 0.077 | ank1a | -0.053 |
| ssr4 | 0.076 | nme2b.2 | -0.053 |
| kazald2 | 0.076 | tnnt3a | -0.052 |
| ubc | 0.073 | acta1b | -0.052 |
| twist1a | 0.073 | glud1b | -0.052 |
| crtap | 0.073 | eno3 | -0.052 |
| si:ch73-335l21.4 | 0.073 | actn3a | -0.052 |
| cd248b | 0.072 | si:ch211-266g18.10 | -0.052 |
| znf503 | 0.072 | pvalb1 | -0.051 |
| btg2 | 0.072 | pgam2 | -0.051 |
| vcana | 0.072 | pvalb2 | -0.051 |
| rrbp1b | 0.071 | smyd1a | -0.051 |
| fgfrl1a | 0.071 | hhatla | -0.051 |
| actb2 | 0.071 | srl | -0.051 |
| ahnak | 0.070 | casq2 | -0.050 |
| calua | 0.069 | hrc | -0.050 |
| arf5 | 0.069 | actn3b | -0.050 |