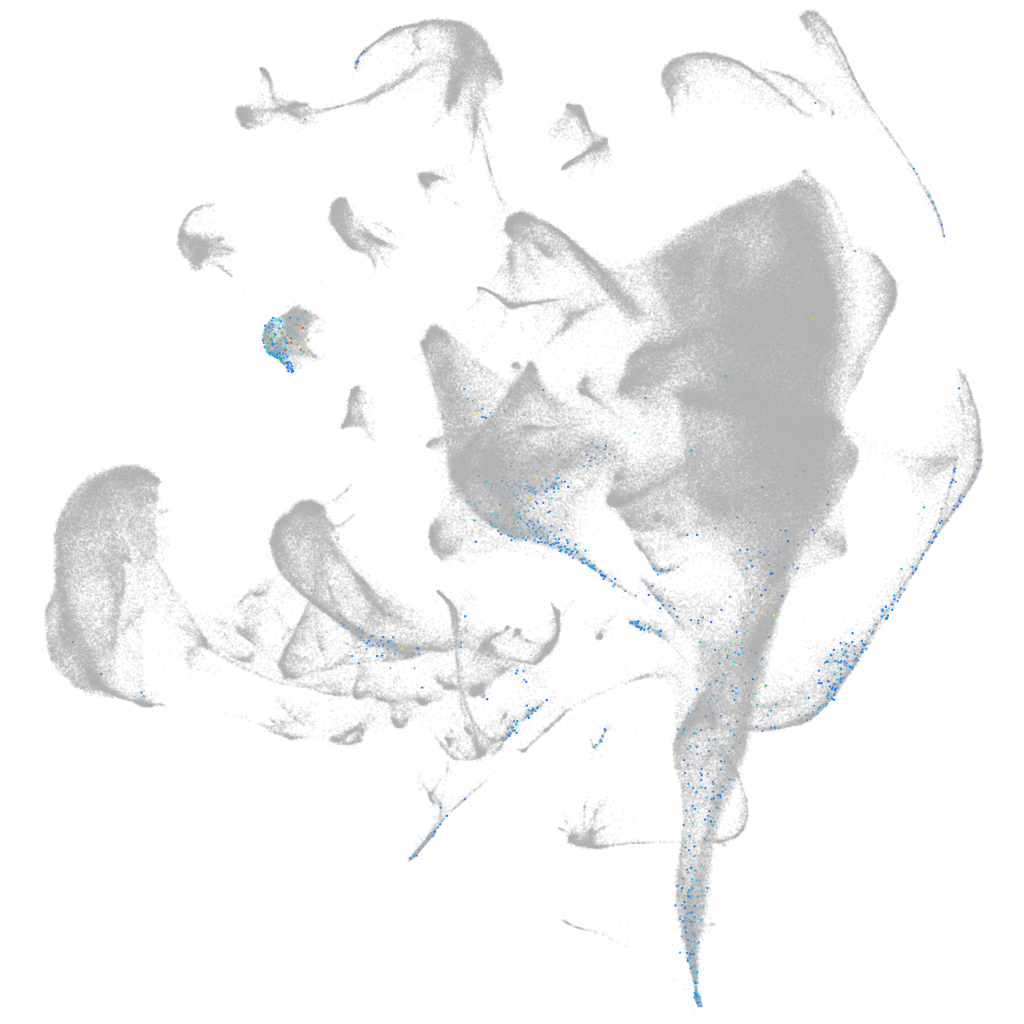
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| etv2 | 0.103 | tuba1c | -0.040 |
| gpr182 | 0.102 | gpm6aa | -0.034 |
| CU570769.1 | 0.100 | elavl3 | -0.031 |
| clec14a | 0.093 | mdkb | -0.031 |
| mrc1a | 0.093 | pvalb1 | -0.031 |
| stab2 | 0.090 | ckbb | -0.030 |
| tnfaip2b | 0.088 | stmn1b | -0.030 |
| sele | 0.087 | rtn1a | -0.029 |
| tie1 | 0.087 | tuba1a | -0.029 |
| egfl7 | 0.085 | pvalb2 | -0.028 |
| hhex | 0.085 | tmsb | -0.028 |
| XLOC-042222 | 0.084 | fabp3 | -0.027 |
| dab2 | 0.081 | fabp7a | -0.027 |
| lyve1b | 0.081 | nova2 | -0.026 |
| cdh5 | 0.080 | rnasekb | -0.026 |
| rasip1 | 0.080 | celf2 | -0.024 |
| scarf1 | 0.080 | cspg5a | -0.024 |
| she | 0.080 | epb41a | -0.024 |
| erg | 0.079 | cadm3 | -0.023 |
| plvapb | 0.075 | CU467822.1 | -0.023 |
| si:dkeyp-97a10.2 | 0.075 | gng3 | -0.023 |
| CT030188.1 | 0.074 | si:ch211-137a8.4 | -0.023 |
| tek | 0.074 | si:dkey-276j7.1 | -0.023 |
| yrk | 0.074 | atp6v0cb | -0.022 |
| pcdh12 | 0.072 | hbbe1.3 | -0.022 |
| tmem88a | 0.072 | icn | -0.022 |
| meox1 | 0.070 | krtt1c19e | -0.022 |
| myct1a | 0.070 | msi1 | -0.022 |
| tpm4a | 0.070 | myt1b | -0.022 |
| ushbp1 | 0.070 | sncb | -0.022 |
| fli1b | 0.069 | fez1 | -0.021 |
| adgrl4 | 0.068 | gnao1a | -0.021 |
| pecam1 | 0.068 | hbae3 | -0.021 |
| ramp2 | 0.068 | icn2 | -0.021 |
| stab1 | 0.068 | krt4 | -0.021 |