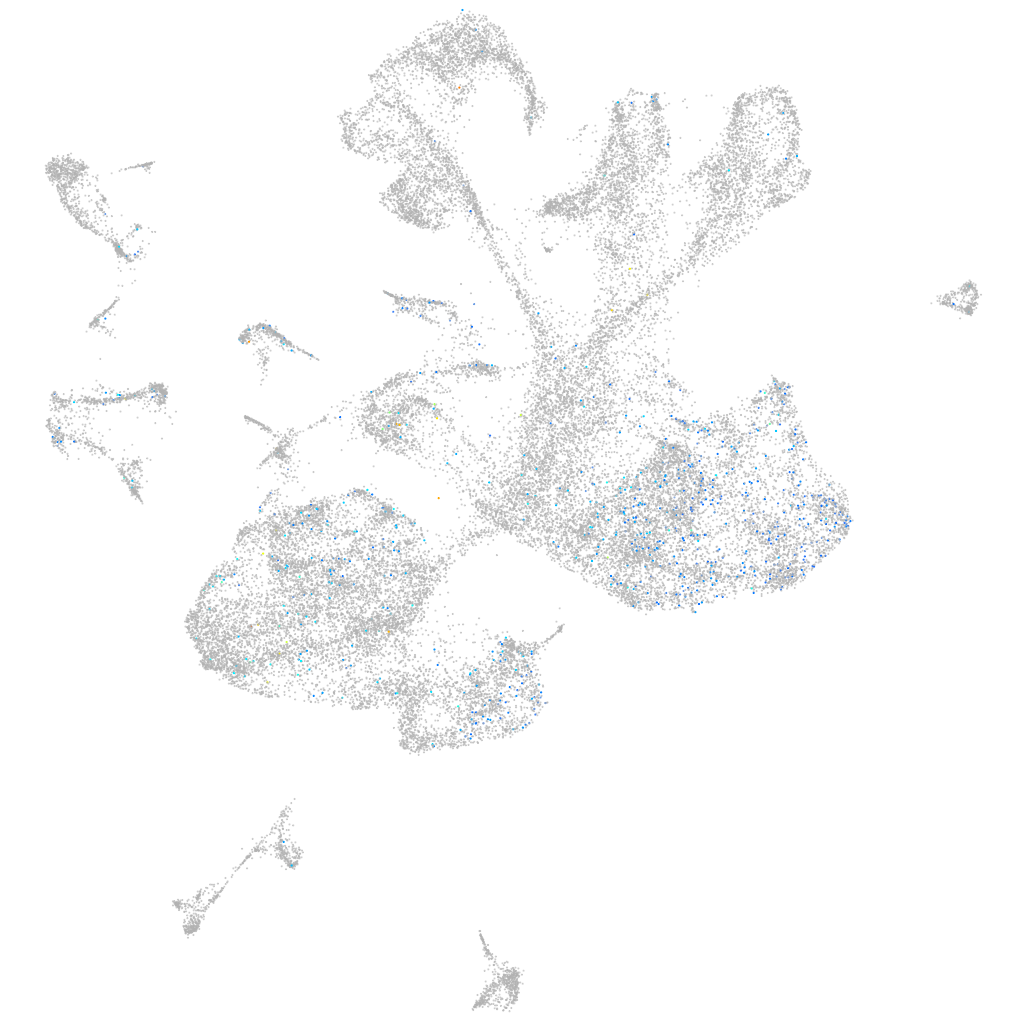
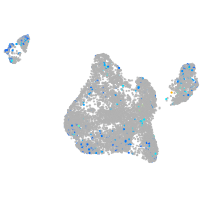
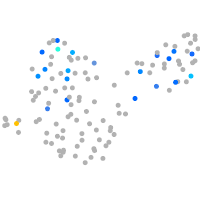
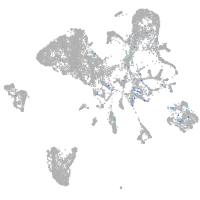
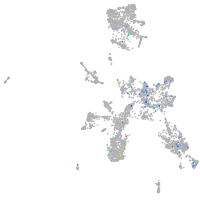
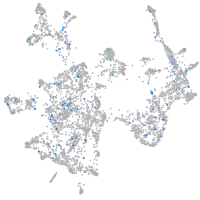
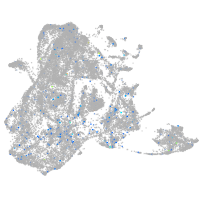
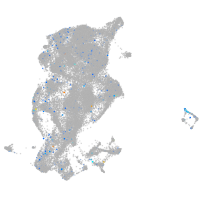
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dut | 0.095 | elavl3 | -0.066 |
| cks1b | 0.093 | rtn1a | -0.059 |
| pcna | 0.091 | stmn1b | -0.054 |
| or133-3 | 0.088 | myt1b | -0.054 |
| mki67 | 0.088 | elavl4 | -0.050 |
| dek | 0.087 | tmsb | -0.049 |
| chaf1a | 0.086 | gng3 | -0.049 |
| ccna2 | 0.086 | ckbb | -0.049 |
| rrm1 | 0.084 | si:dkeyp-75h12.5 | -0.049 |
| smc2 | 0.083 | calm1a | -0.047 |
| aurkb | 0.083 | marcksl1b | -0.047 |
| mibp | 0.082 | rnasekb | -0.046 |
| LOC100330864 | 0.081 | rtn1b | -0.045 |
| zgc:110540 | 0.081 | gpm6ab | -0.045 |
| rrm2 | 0.080 | stx1b | -0.045 |
| anp32b | 0.080 | scrt2 | -0.045 |
| banf1 | 0.079 | atp6v0cb | -0.044 |
| tuba8l4 | 0.079 | stxbp1a | -0.044 |
| CABZ01005379.1 | 0.079 | ywhah | -0.044 |
| tuba8l | 0.079 | sncb | -0.044 |
| stmn1a | 0.079 | syt2a | -0.043 |
| hmgb2a | 0.079 | tubb5 | -0.042 |
| cdca5 | 0.078 | dpysl3 | -0.041 |
| selenoh | 0.078 | gng2 | -0.041 |
| lbr | 0.077 | nsg2 | -0.041 |
| fen1 | 0.076 | cplx2 | -0.041 |
| esco2 | 0.076 | myt1a | -0.041 |
| dhfr | 0.076 | LOC100537384 | -0.041 |
| rpa2 | 0.076 | vamp2 | -0.041 |
| mad2l1 | 0.075 | map1aa | -0.041 |
| nutf2l | 0.075 | onecut2 | -0.041 |
| ncapg | 0.074 | ube2d4 | -0.040 |
| haus4 | 0.073 | zc4h2 | -0.040 |
| fbxo5 | 0.073 | snap25a | -0.040 |
| suv39h1b | 0.072 | gap43 | -0.040 |