Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme 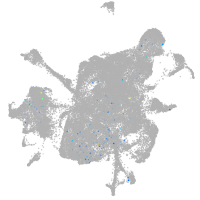 |
hematopoietic / vasculature 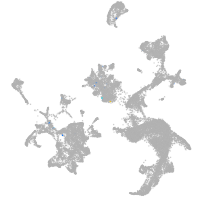 |
pronephros  |
neural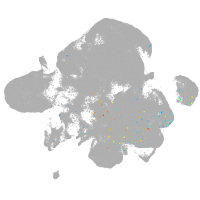 |
spinal cord / glia |
eye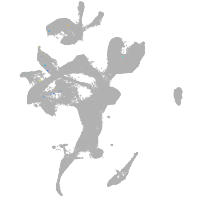 |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| fkbp6 | 0.195 | ttn.1 | -0.043 |
| cd248b | 0.118 | cycsb | -0.041 |
| igf2b | 0.117 | hsp90aa1.1 | -0.040 |
| CU694486.1 | 0.111 | ttn.2 | -0.039 |
| nkx3-1 | 0.109 | actc1b | -0.039 |
| si:dkey-43f9.4 | 0.107 | atp5f1e | -0.039 |
| si:ch211-286o17.1 | 0.102 | cox7c | -0.038 |
| meis3 | 0.102 | atp5if1b | -0.036 |
| arhgap36 | 0.102 | atp2a1 | -0.035 |
| tmem88b | 0.097 | klhl41b | -0.035 |
| inka1a | 0.097 | ckmb | -0.035 |
| colec12 | 0.096 | ak1 | -0.035 |
| CU694807.1 | 0.095 | gapdh | -0.034 |
| irx7 | 0.094 | fxr2 | -0.034 |
| foxp1b | 0.094 | tmem38a | -0.034 |
| foxd1 | 0.093 | aldoab | -0.034 |
| nkx3.2 | 0.093 | chchd10 | -0.034 |
| lin28a | 0.093 | ckma | -0.034 |
| ets1 | 0.093 | zgc:92429 | -0.033 |
| twist1b | 0.093 | pabpc4 | -0.033 |
| LOC101883267 | 0.092 | tnnc2 | -0.033 |
| foxd2 | 0.090 | desma | -0.033 |
| CU571069.1 | 0.089 | cox17 | -0.033 |
| col11a1a | 0.089 | mybphb | -0.032 |
| zfp36l2 | 0.088 | acta1b | -0.032 |
| tpm4a | 0.088 | gatm | -0.032 |
| LO018188.1 | 0.087 | rbfox1l | -0.032 |
| angptl7 | 0.087 | tpma | -0.032 |
| irf2bp2b | 0.086 | nme2b.2 | -0.032 |
| krt18a.1 | 0.084 | eno3 | -0.032 |
| kirrel1b | 0.084 | tob1a | -0.031 |
| sntg2 | 0.083 | neb | -0.031 |
| rbp5 | 0.082 | zgc:101853 | -0.031 |
| fibina | 0.081 | acta1a | -0.031 |
| sdc4 | 0.079 | ldb3a | -0.031 |




