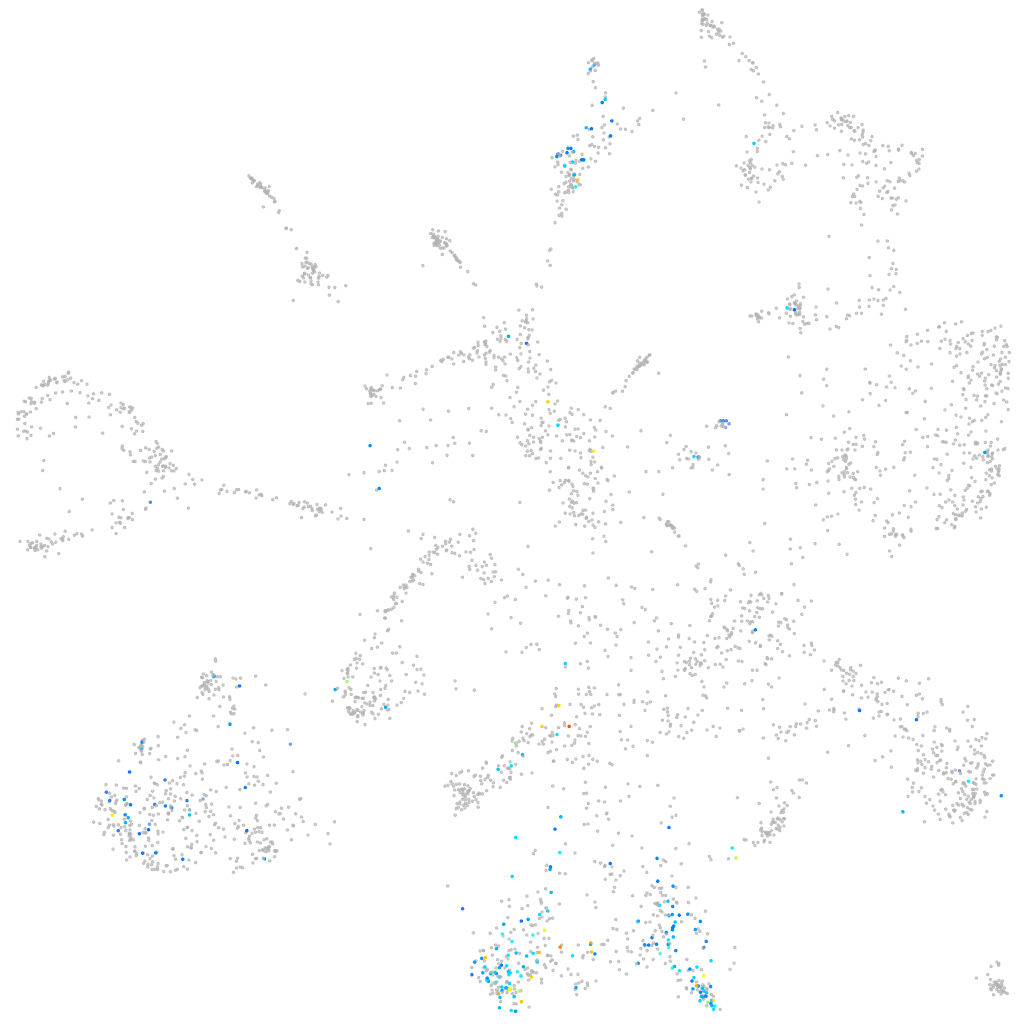
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle 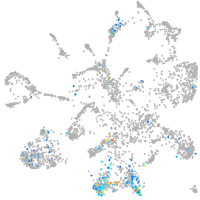 |
mesenchyme 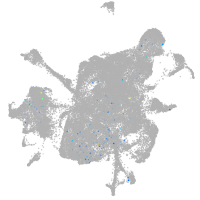 |
hematopoietic / vasculature 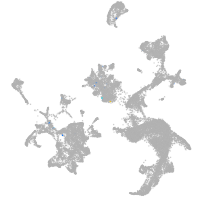 |
pronephros  |
neural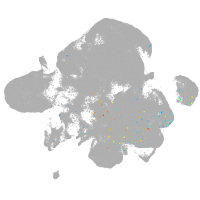 |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cald1b | 0.395 | tmem88b | -0.118 |
| desmb | 0.394 | marcksl1b | -0.113 |
| si:ch211-137i24.10 | 0.375 | fthl27 | -0.110 |
| cnn1b | 0.369 | jam2b | -0.110 |
| myh11a | 0.347 | podxl | -0.110 |
| tpm2 | 0.346 | nme2b.1 | -0.102 |
| csrp1b | 0.344 | ptmab | -0.099 |
| fhl1a | 0.319 | h3f3a | -0.096 |
| tagln | 0.302 | pmp22a | -0.096 |
| cabp1a | 0.299 | cpn1 | -0.094 |
| vcla | 0.297 | sparc | -0.093 |
| mylkb | 0.297 | bambia | -0.092 |
| fbxl22 | 0.296 | ctsla | -0.091 |
| acta2 | 0.295 | hmga1a | -0.089 |
| myl6 | 0.292 | fgfr2 | -0.088 |
| actc1a | 0.285 | rpl38 | -0.087 |
| myl9a | 0.284 | ednraa | -0.081 |
| arhgap20b | 0.279 | rbpms2b | -0.080 |
| itpka | 0.278 | si:ch211-160j14.3 | -0.080 |
| tpm1 | 0.275 | krt94 | -0.079 |
| zgc:110699 | 0.274 | bcam | -0.079 |
| fxyd1 | 0.271 | h3f3d | -0.078 |
| smtnb | 0.262 | si:ch73-281n10.2 | -0.078 |
| flnca | 0.260 | barx1 | -0.077 |
| CR753876.1 | 0.259 | her9 | -0.077 |
| CU571069.1 | 0.259 | pdgfra | -0.076 |
| acta1b | 0.256 | fabp11a | -0.076 |
| rgs2 | 0.254 | raraa | -0.076 |
| rbm24a | 0.250 | aqp1a.1 | -0.075 |
| pnp4a | 0.248 | si:ch211-39i22.1 | -0.075 |
| si:ch211-62a1.3 | 0.247 | mfap5 | -0.074 |
| ckbb | 0.245 | hmgb2a | -0.074 |
| si:dkeyp-51f12.2 | 0.244 | cirbpb | -0.073 |
| ckba | 0.243 | ackr3b | -0.073 |
| pdlim3a | 0.241 | stmn1a | -0.073 |