kringle containing transmembrane protein 1
ZFIN 
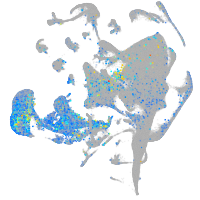
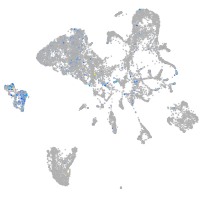
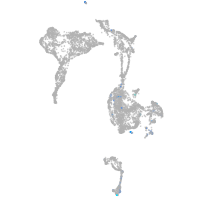
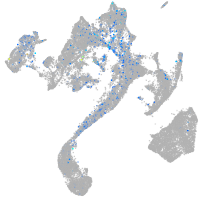
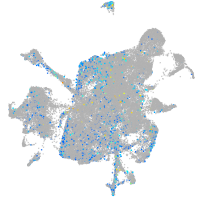
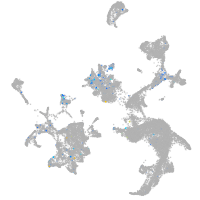
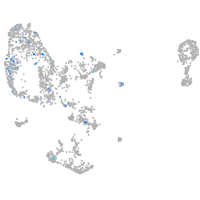
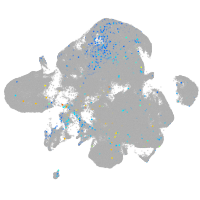
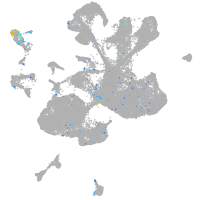
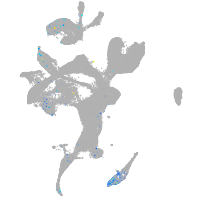
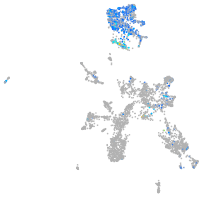
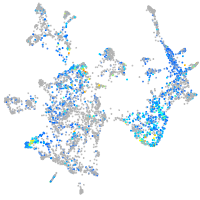
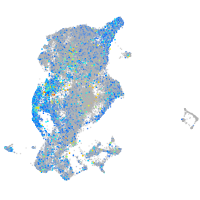
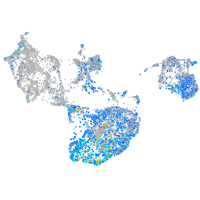
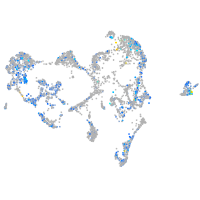
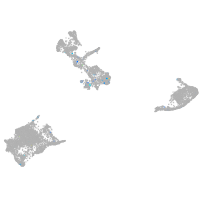
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ifitm5 | 0.187 | fgfbp2b | -0.073 |
| tmem119a | 0.182 | cnmd | -0.067 |
| panx3 | 0.181 | col9a2 | -0.062 |
| phospho1 | 0.176 | col2a1a | -0.061 |
| sgms2a | 0.173 | col9a3 | -0.058 |
| col10a1a | 0.170 | col9a1a | -0.058 |
| col5a2b | 0.169 | sox9a | -0.050 |
| slc8a4b | 0.166 | foxc1b | -0.048 |
| cd81b | 0.163 | otos | -0.048 |
| si:ch211-106n13.3 | 0.149 | matn1 | -0.046 |
| phex | 0.149 | mia | -0.044 |
| entpd5a | 0.146 | acana | -0.043 |
| enam | 0.145 | matn4 | -0.042 |
| bgna | 0.144 | epyc | -0.041 |
| pltp | 0.141 | ucmab | -0.041 |
| si:dkey-32e6.6 | 0.130 | snorc | -0.041 |
| smpd3 | 0.129 | ebf3a | -0.041 |
| sp7 | 0.122 | AL929222.2 | -0.041 |
| sod3b | 0.122 | cxcl12a | -0.040 |
| ano5a | 0.121 | zgc:162879 | -0.040 |
| tcf7 | 0.121 | tnxba | -0.040 |
| myadml2 | 0.120 | ptgdsa | -0.039 |
| bmp8a | 0.112 | wisp3 | -0.038 |
| zbtb7c | 0.106 | trabd2a | -0.038 |
| spp1 | 0.106 | si:ch211-251b21.1 | -0.037 |
| mfsd2b | 0.102 | wwp2 | -0.037 |
| notum1a | 0.099 | sept8b | -0.036 |
| ppfibp2a | 0.096 | si:dkey-19b23.8 | -0.036 |
| scpp1 | 0.092 | VIT | -0.035 |
| zgc:92162 | 0.088 | si:dkey-6n6.1 | -0.035 |
| si:cabz01100188.1 | 0.087 | emilin3a | -0.035 |
| dlx3b | 0.086 | pcolceb | -0.035 |
| cadm1b | 0.085 | sox5 | -0.034 |
| chst3b | 0.084 | prelp | -0.034 |
| lamc3 | 0.084 | nog3 | -0.033 |