karyopherin (importin) beta 1
ZFIN 
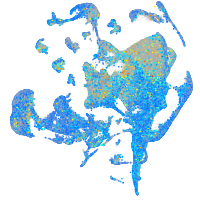
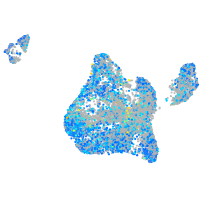
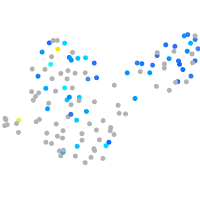
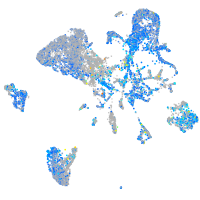
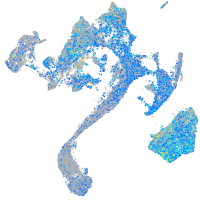
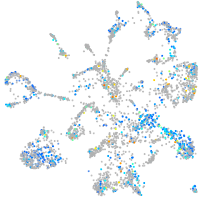
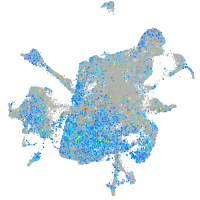
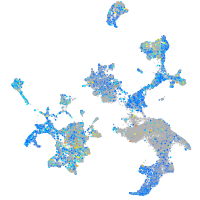
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pcna | 0.322 | actc1b | -0.249 |
| anp32b | 0.313 | aldoab | -0.224 |
| seta | 0.306 | ak1 | -0.222 |
| tuba8l4 | 0.301 | atp2a1 | -0.219 |
| hmgb2a | 0.299 | ttn.2 | -0.217 |
| chaf1a | 0.298 | acta1b | -0.217 |
| ppm1g | 0.295 | tnnc2 | -0.217 |
| stmn1a | 0.293 | tmem38a | -0.215 |
| hnrnpabb | 0.292 | ckmb | -0.215 |
| slbp | 0.291 | tpma | -0.214 |
| nasp | 0.290 | ckma | -0.213 |
| si:ch211-222l21.1 | 0.290 | ttn.1 | -0.213 |
| banf1 | 0.290 | mybphb | -0.208 |
| cirbpa | 0.289 | srl | -0.207 |
| sumo3b | 0.287 | acta1a | -0.206 |
| cbx3a | 0.287 | desma | -0.204 |
| hmga1a | 0.286 | ldb3a | -0.204 |
| hmgb2b | 0.282 | neb | -0.203 |
| hnrnpa1b | 0.280 | mylpfa | -0.203 |
| anp32a | 0.279 | CABZ01078594.1 | -0.202 |
| tubb2b | 0.279 | cav3 | -0.200 |
| hnrnpa0b | 0.278 | ldb3b | -0.193 |
| lig1 | 0.277 | eno1a | -0.192 |
| dek | 0.276 | casq2 | -0.190 |
| si:ch211-288g17.3 | 0.275 | gapdh | -0.190 |
| rbbp4 | 0.275 | actn3b | -0.190 |
| baz1b | 0.275 | CABZ01072309.1 | -0.188 |
| mcm7 | 0.273 | tnnt3a | -0.188 |
| ranbp1 | 0.273 | gamt | -0.187 |
| rrm1 | 0.273 | nme2b.2 | -0.187 |
| khdrbs1a | 0.271 | myl1 | -0.186 |
| taf15 | 0.271 | actc1a | -0.185 |
| sumo3a | 0.269 | si:ch73-367p23.2 | -0.185 |
| setb | 0.268 | pabpc4 | -0.184 |
| hnrnpaba | 0.268 | txlnbb | -0.184 |