Kruppel-like factor 7b
ZFIN 
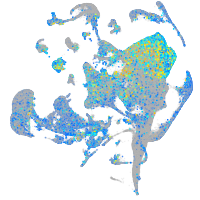
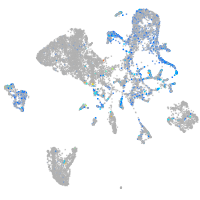
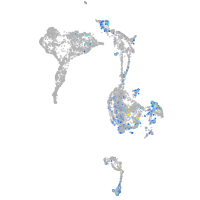
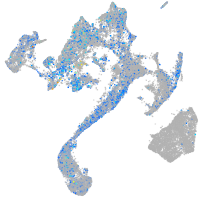
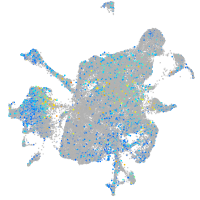
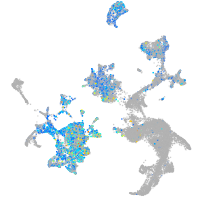
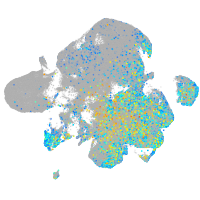
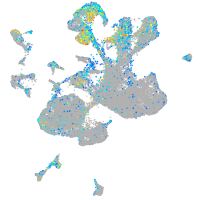
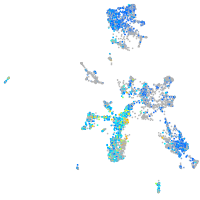
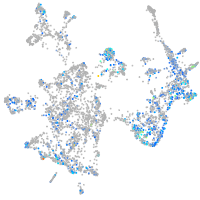
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cldnh | 0.295 | bhmt | -0.250 |
| cap1 | 0.271 | gamt | -0.239 |
| icn | 0.270 | gatm | -0.232 |
| ier2b | 0.267 | aqp12 | -0.227 |
| si:ch211-137i24.10 | 0.262 | mat1a | -0.220 |
| phlda2 | 0.256 | agxtb | -0.220 |
| epcam | 0.254 | apoc1 | -0.203 |
| elovl1b | 0.250 | kng1 | -0.200 |
| calm1b | 0.248 | ttc36 | -0.198 |
| tagln2 | 0.245 | tfa | -0.197 |
| capns1b | 0.244 | grhprb | -0.197 |
| pfn1 | 0.242 | apoa1b | -0.194 |
| cd63 | 0.241 | serpina1l | -0.193 |
| krt4 | 0.241 | serpina1 | -0.193 |
| si:ch1073-429i10.3.1 | 0.239 | hao1 | -0.193 |
| actb1 | 0.238 | rbp2b | -0.192 |
| degs2 | 0.238 | fgg | -0.191 |
| si:dkeyp-72e1.6 | 0.236 | ahcy | -0.191 |
| CR383676.1 | 0.235 | fabp10a | -0.191 |
| zgc:92380 | 0.235 | apom | -0.191 |
| gnb1a | 0.234 | pnp4b | -0.190 |
| spint2 | 0.234 | ambp | -0.190 |
| prr15la | 0.233 | apoa2 | -0.190 |
| cldnb | 0.233 | uox | -0.189 |
| calm1a | 0.232 | hpda | -0.188 |
| tmem45b | 0.231 | zgc:123103 | -0.187 |
| mcl1a | 0.231 | ces2 | -0.185 |
| mvp | 0.230 | ttr | -0.185 |
| ftr83 | 0.230 | fetub | -0.185 |
| capn2b | 0.230 | fgb | -0.185 |
| nipa2 | 0.230 | f2 | -0.184 |
| sri | 0.229 | LOC110437731 | -0.184 |
| ubc | 0.228 | vtnb | -0.182 |
| f11r.1 | 0.228 | serpinf2b | -0.181 |
| serpinb1 | 0.228 | si:dkey-16p21.8 | -0.181 |