kinesin light chain 1a
ZFIN 
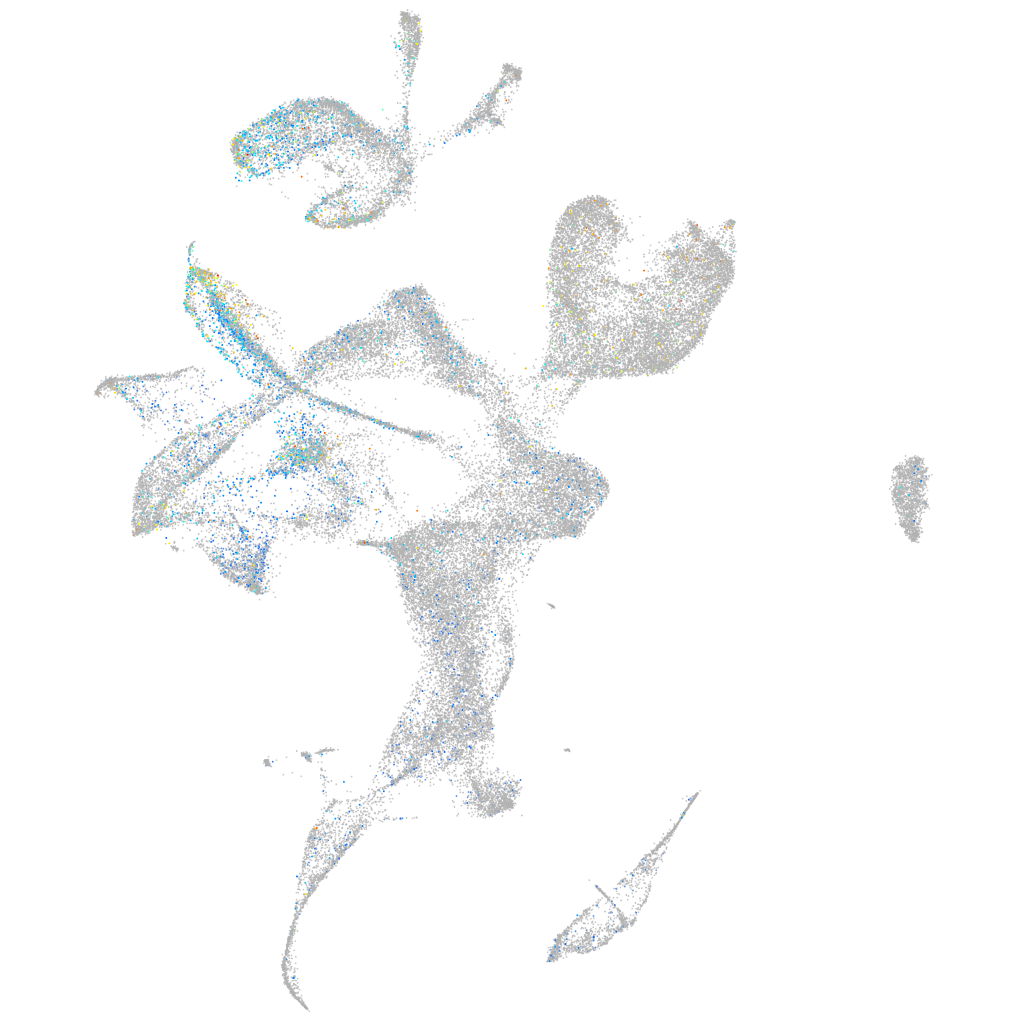
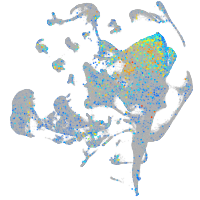
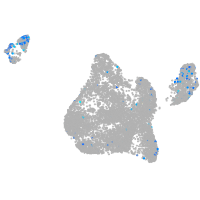
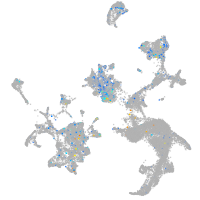
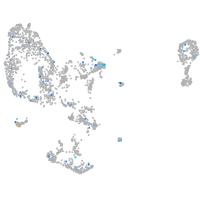
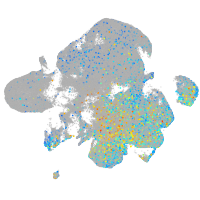
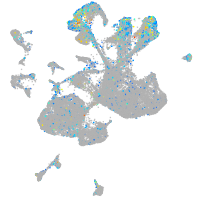
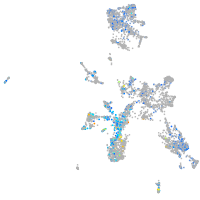
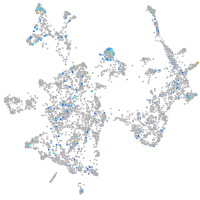
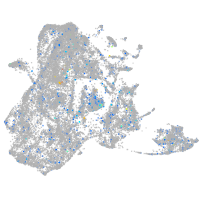
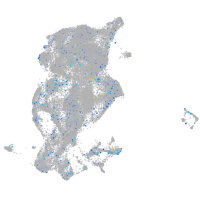
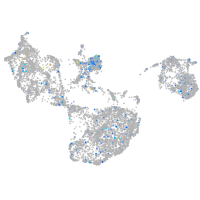
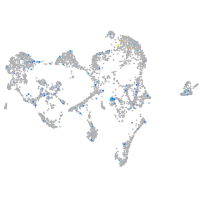
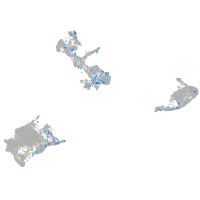
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| eno2 | 0.200 | hmgb2a | -0.135 |
| stmn2a | 0.198 | pcna | -0.083 |
| rtn1b | 0.186 | hmga1a | -0.082 |
| zgc:65894 | 0.186 | rrm1 | -0.079 |
| sncb | 0.185 | stmn1a | -0.078 |
| stxbp1a | 0.184 | ahcy | -0.077 |
| ywhag2 | 0.183 | lbr | -0.075 |
| rbpms2a | 0.182 | mdka | -0.075 |
| snap25a | 0.180 | mki67 | -0.075 |
| elavl3 | 0.177 | mcm7 | -0.074 |
| stx1b | 0.177 | ccnd1 | -0.074 |
| syt2a | 0.176 | ccna2 | -0.073 |
| zgc:153426 | 0.173 | nutf2l | -0.073 |
| elavl4 | 0.173 | selenoh | -0.072 |
| sv2a | 0.172 | dut | -0.071 |
| cplx2l | 0.169 | her15.1 | -0.071 |
| id4 | 0.168 | msna | -0.071 |
| gng3 | 0.168 | chaf1a | -0.069 |
| vsnl1b | 0.167 | anp32b | -0.069 |
| inab | 0.165 | hmgb2b | -0.068 |
| maptb | 0.162 | tuba8l | -0.067 |
| islr2 | 0.161 | lig1 | -0.067 |
| tkta | 0.159 | mcm6 | -0.066 |
| gap43 | 0.158 | banf1 | -0.066 |
| rbpms2b | 0.158 | cx43.4 | -0.066 |
| gnao1a | 0.157 | fen1 | -0.065 |
| tuba2 | 0.157 | id1 | -0.065 |
| ppp1r14ba | 0.155 | rpa3 | -0.064 |
| isl2b | 0.155 | dlgap5 | -0.064 |
| mllt11 | 0.154 | COX7A2 (1 of many) | -0.064 |
| cdk5r1b | 0.154 | nasp | -0.063 |
| syt1a | 0.152 | dek | -0.063 |
| stmn1b | 0.151 | CABZ01005379.1 | -0.063 |
| sprn | 0.151 | cdca7b | -0.063 |
| pou4f1 | 0.151 | cks1b | -0.063 |