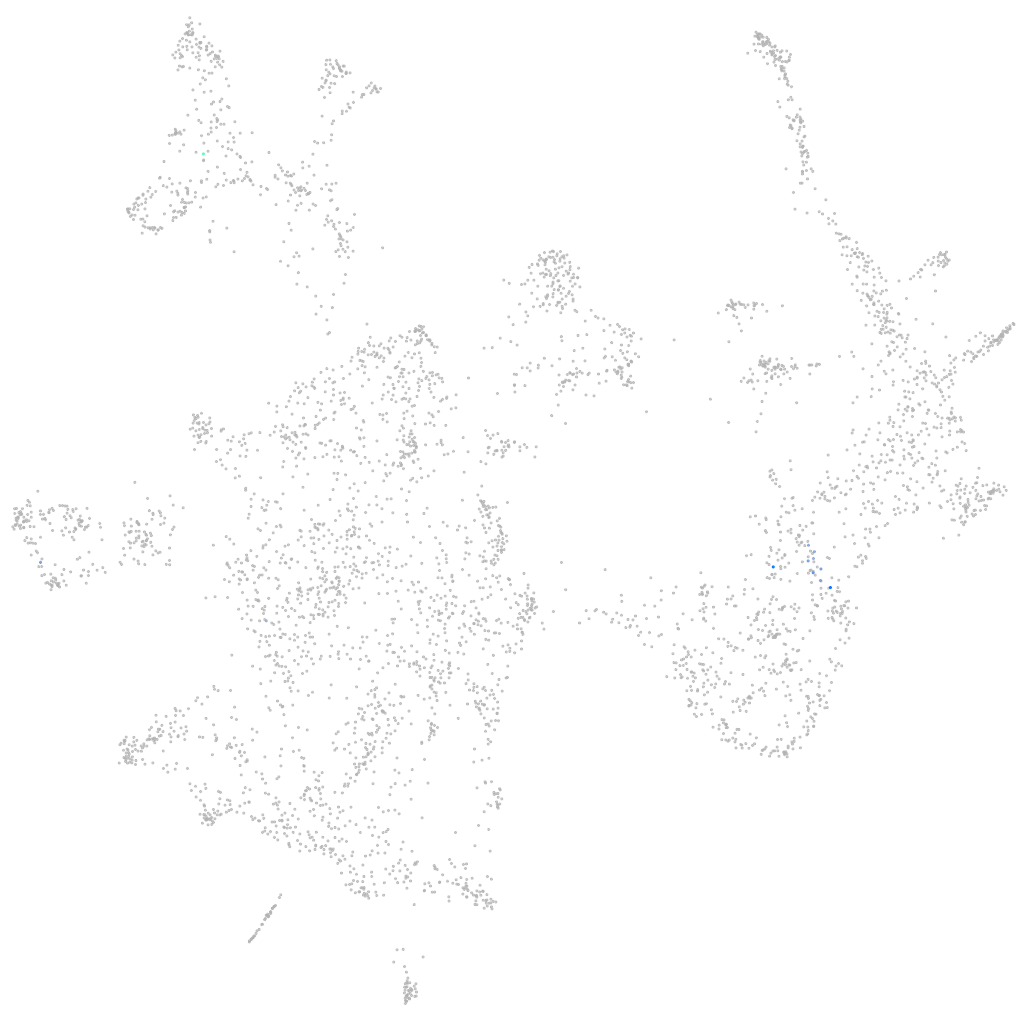
KiSS-1 metastasis suppressor
ZFIN 
Other cell groups


all cells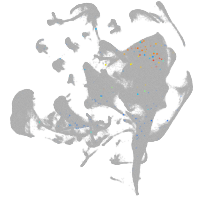 |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 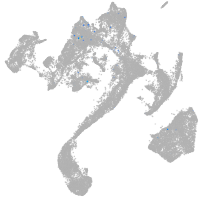 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cdc42ep3 | 0.449 | rpl31 | -0.054 |
| chrnb3a | 0.376 | rps27a | -0.049 |
| steap2 | 0.360 | rps14 | -0.048 |
| dre-let-7i | 0.353 | rpl21 | -0.046 |
| rhobtb1 | 0.310 | rpl36 | -0.044 |
| zgc:153431 | 0.279 | CR383676.1 | -0.043 |
| acot7 | 0.278 | rpl18a | -0.042 |
| slc12a7b | 0.264 | rpl18 | -0.042 |
| tent5c | 0.234 | sec61g | -0.041 |
| trim32 | 0.229 | rpl35a | -0.041 |
| XLOC-043073 | 0.227 | rack1 | -0.041 |
| CR388166.2 | 0.222 | atp1b1a | -0.040 |
| asb3 | 0.217 | rpl24 | -0.039 |
| adcy6a | 0.214 | mt-atp6 | -0.039 |
| si:dkey-32e6.3 | 0.208 | pnrc2 | -0.039 |
| si:ch73-285p12.4 | 0.206 | rps28 | -0.038 |
| shisa3 | 0.206 | fthl27 | -0.038 |
| XLOC-030237 | 0.205 | itm2ba | -0.037 |
| tm7sf2 | 0.205 | eef1da | -0.036 |
| polr3a | 0.200 | stm | -0.036 |
| bmp2k | 0.197 | rps26 | -0.034 |
| tubgcp5 | 0.195 | ctsla | -0.034 |
| kcnk1b | 0.195 | rpl38 | -0.034 |
| tyw5 | 0.194 | eif1b | -0.033 |
| pla2g4ab | 0.190 | rpl3 | -0.033 |
| arhgap33 | 0.186 | id2a | -0.032 |
| pou3f2b | 0.180 | sparc | -0.032 |
| pcolce2b | 0.177 | rps3 | -0.031 |
| zgc:154075 | 0.175 | rps27.2 | -0.031 |
| them4 | 0.171 | naca | -0.031 |
| wdr95 | 0.170 | rps12 | -0.031 |
| si:dkey-1h24.2 | 0.170 | col2a1a | -0.031 |
| alkbh7 | 0.168 | appa | -0.031 |
| hspa13 | 0.168 | spint2 | -0.030 |
| impact | 0.167 | agr2 | -0.030 |