kirre like nephrin family adhesion molecule 3a
ZFIN 
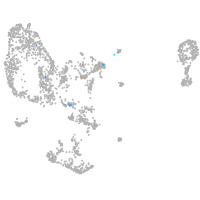
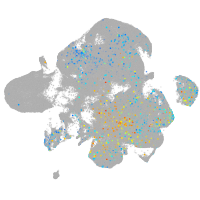
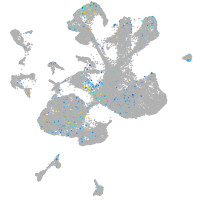
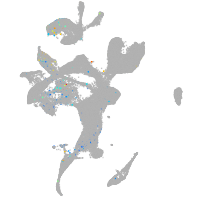
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| eno2 | 0.128 | hmgb2a | -0.086 |
| sv2a | 0.124 | hmga1a | -0.046 |
| tkta | 0.124 | lbr | -0.044 |
| rtn1b | 0.120 | rrm1 | -0.044 |
| stx1b | 0.114 | ahcy | -0.041 |
| stxbp1a | 0.114 | nutf2l | -0.041 |
| sprn | 0.112 | fabp7a | -0.040 |
| vsnl1b | 0.112 | hmgb2b | -0.040 |
| elavl4 | 0.112 | neurod4 | -0.039 |
| elavl3 | 0.112 | stmn1a | -0.039 |
| snap25a | 0.111 | ccna2 | -0.039 |
| rbpms2a | 0.111 | dek | -0.039 |
| ywhag2 | 0.111 | pcna | -0.039 |
| slc35g2b | 0.110 | mki67 | -0.039 |
| myt1la | 0.108 | anp32e | -0.037 |
| stmn2a | 0.108 | zgc:110425 | -0.037 |
| syt2a | 0.108 | msi1 | -0.037 |
| syt1a | 0.105 | msna | -0.037 |
| cplx2l | 0.105 | dut | -0.036 |
| zgc:153426 | 0.102 | crx | -0.036 |
| gabrb4 | 0.101 | chaf1a | -0.036 |
| pou4f3 | 0.100 | zgc:110216 | -0.036 |
| zgc:65894 | 0.099 | cks1b | -0.036 |
| zgc:171482 | 0.099 | rrm2 | -0.036 |
| ywhag1 | 0.098 | mibp | -0.035 |
| gng3 | 0.098 | ddah2 | -0.035 |
| nrxn2a | 0.097 | ccnd1 | -0.035 |
| scn8aa | 0.097 | tuba8l4 | -0.034 |
| sncb | 0.097 | otx5 | -0.034 |
| glra4b | 0.096 | her15.1 | -0.034 |
| anks1ab | 0.096 | tuba8l | -0.033 |
| ppp1r14ba | 0.095 | rps20 | -0.033 |
| IGLON5 | 0.095 | rpl11 | -0.033 |
| nrn1a | 0.095 | ncapg | -0.033 |
| olfm2a | 0.094 | rpa3 | -0.033 |