kinesin family-like
ZFIN 
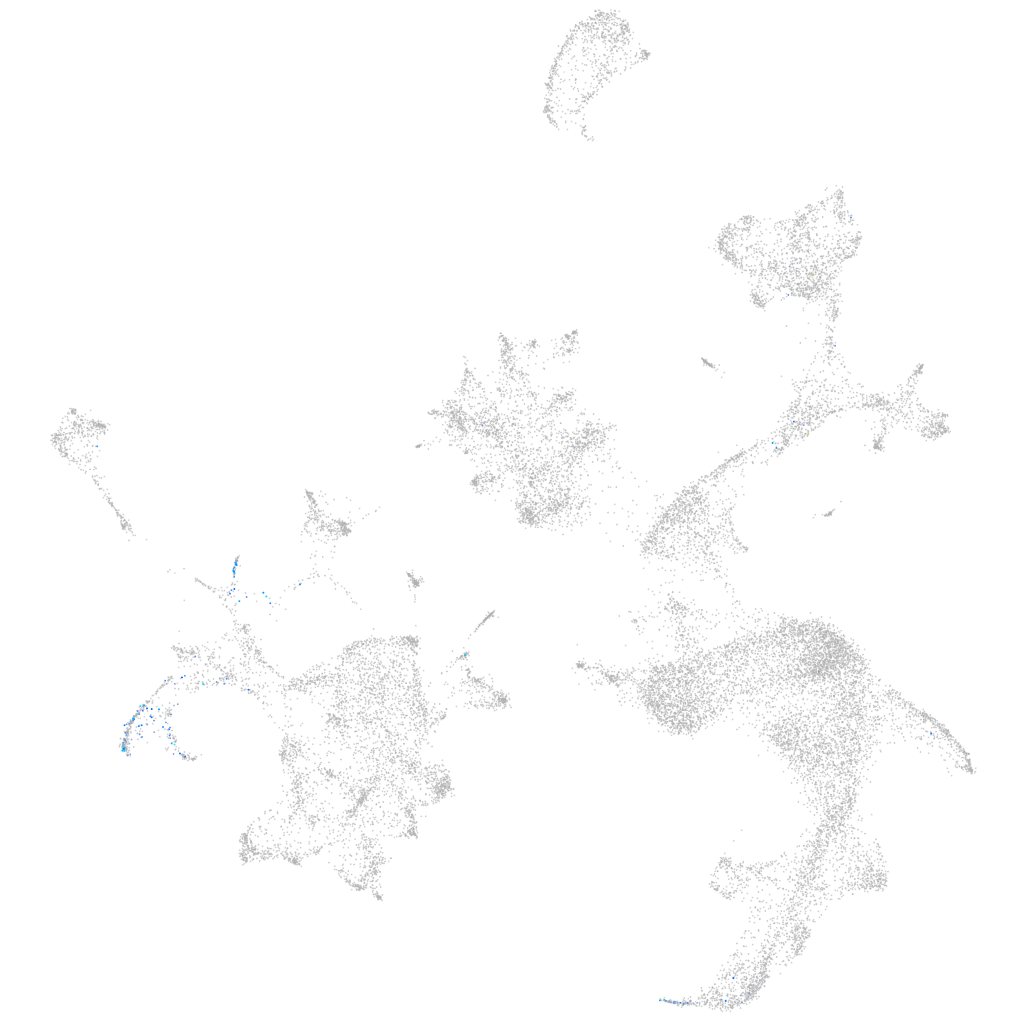
Other cell groups


all cells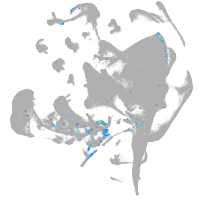 |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 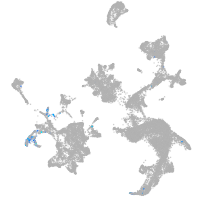 |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin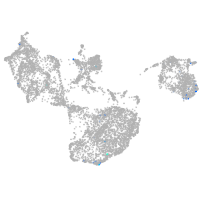 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| morc3b | 0.258 | pvalb1 | -0.054 |
| wu:fb97g03 | 0.240 | pvalb2 | -0.050 |
| si:dkey-261j4.4 | 0.236 | actc1b | -0.047 |
| gfi1aa | 0.210 | gpx1a | -0.047 |
| zgc:123297 | 0.203 | ccni | -0.043 |
| tcf21 | 0.201 | si:ch211-250g4.3 | -0.041 |
| si:ch73-299h12.2 | 0.199 | tmod4 | -0.041 |
| si:dkey-261j4.3 | 0.185 | hbbe1.3 | -0.040 |
| igfbp1b | 0.184 | gapdhs | -0.038 |
| ifit10 | 0.178 | cahz | -0.037 |
| cldng | 0.177 | plac8l1 | -0.036 |
| hspb8 | 0.174 | hbbe2 | -0.036 |
| inka1b | 0.166 | sod1 | -0.036 |
| drl | 0.166 | srgn | -0.036 |
| mycb | 0.165 | slc4a1a | -0.035 |
| setx | 0.161 | tspo | -0.035 |
| hs3st3b1a | 0.160 | bloc1s6 | -0.035 |
| BX005445.1 | 0.158 | mylpfa | -0.035 |
| nifk | 0.155 | gabarapa | -0.033 |
| cebpa | 0.152 | hbbe1.2 | -0.033 |
| klf17 | 0.151 | creg1 | -0.033 |
| myb | 0.149 | si:dkey-240n22.3 | -0.033 |
| rcor2 | 0.147 | hbbe1.1 | -0.033 |
| blf | 0.146 | ak1 | -0.032 |
| ncs1a | 0.145 | ndrg2 | -0.032 |
| cpox | 0.144 | mylz3 | -0.031 |
| pprc1 | 0.143 | aqp1a.1 | -0.031 |
| emg1 | 0.143 | tnni2a.4 | -0.030 |
| si:dkey-44g23.5 | 0.140 | si:ch211-260e23.9 | -0.030 |
| tomm20a | 0.139 | myhz1.1 | -0.030 |
| tma16 | 0.139 | vat1 | -0.030 |
| LOC108179803 | 0.138 | rhag | -0.029 |
| tomm40l | 0.138 | cpn1 | -0.029 |
| LOC100535857 | 0.138 | blvrb | -0.029 |
| rpl7l1 | 0.138 | zgc:163080 | -0.028 |