kinesin family member 7
ZFIN 
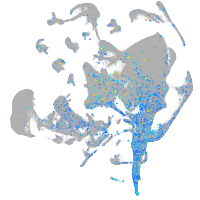
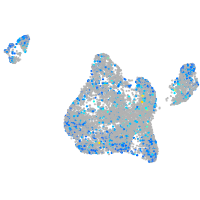
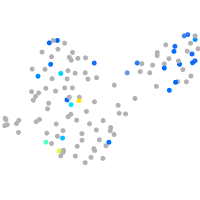
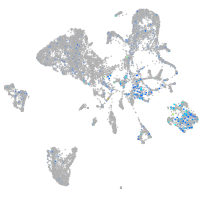
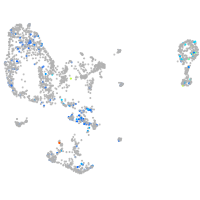
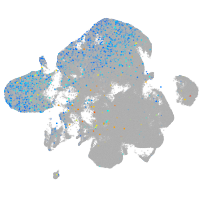
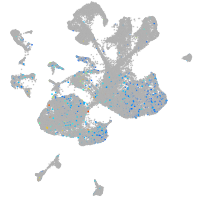
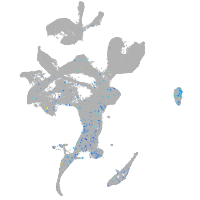
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| id1 | 0.092 | elavl3 | -0.086 |
| XLOC-003690 | 0.081 | myt1b | -0.080 |
| cldn5a | 0.081 | stmn1b | -0.080 |
| tuba8l | 0.079 | gng3 | -0.078 |
| notch3 | 0.078 | rtn1a | -0.073 |
| msna | 0.075 | stxbp1a | -0.071 |
| hmgb2a | 0.074 | elavl4 | -0.070 |
| mki67 | 0.070 | stx1b | -0.070 |
| vamp3 | 0.070 | snap25a | -0.068 |
| akap12b | 0.070 | vamp2 | -0.067 |
| tuba8l4 | 0.069 | atp6v0cb | -0.067 |
| sdc4 | 0.069 | sncb | -0.066 |
| banf1 | 0.067 | tmem59l | -0.065 |
| sox2 | 0.067 | cplx2 | -0.063 |
| her15.1 | 0.066 | rtn1b | -0.063 |
| sox19a | 0.066 | jagn1a | -0.063 |
| zfp36l1a | 0.064 | onecut1 | -0.063 |
| fosab | 0.064 | stmn2a | -0.063 |
| si:ch211-212k18.5 | 0.063 | ptmaa | -0.062 |
| msi1 | 0.062 | nsg2 | -0.062 |
| si:ch73-281n10.2 | 0.062 | si:dkeyp-75h12.5 | -0.062 |
| smc2 | 0.061 | tmsb | -0.062 |
| XLOC-003689 | 0.061 | calm1a | -0.061 |
| wls | 0.061 | golga7ba | -0.061 |
| sox3 | 0.061 | gap43 | -0.060 |
| ptprfb | 0.061 | gpm6ab | -0.060 |
| smo | 0.060 | gng2 | -0.060 |
| mdka | 0.060 | syt2a | -0.060 |
| COX7A2 (1 of many) | 0.060 | scrt2 | -0.060 |
| her2 | 0.060 | rbfox1 | -0.059 |
| cd82a | 0.059 | zc4h2 | -0.058 |
| boc | 0.059 | mllt11 | -0.058 |
| ptbp1a | 0.059 | rnasekb | -0.058 |
| ccnb1 | 0.058 | cplx2l | -0.058 |
| dut | 0.058 | ywhah | -0.057 |