kinesin family member 21B
ZFIN 
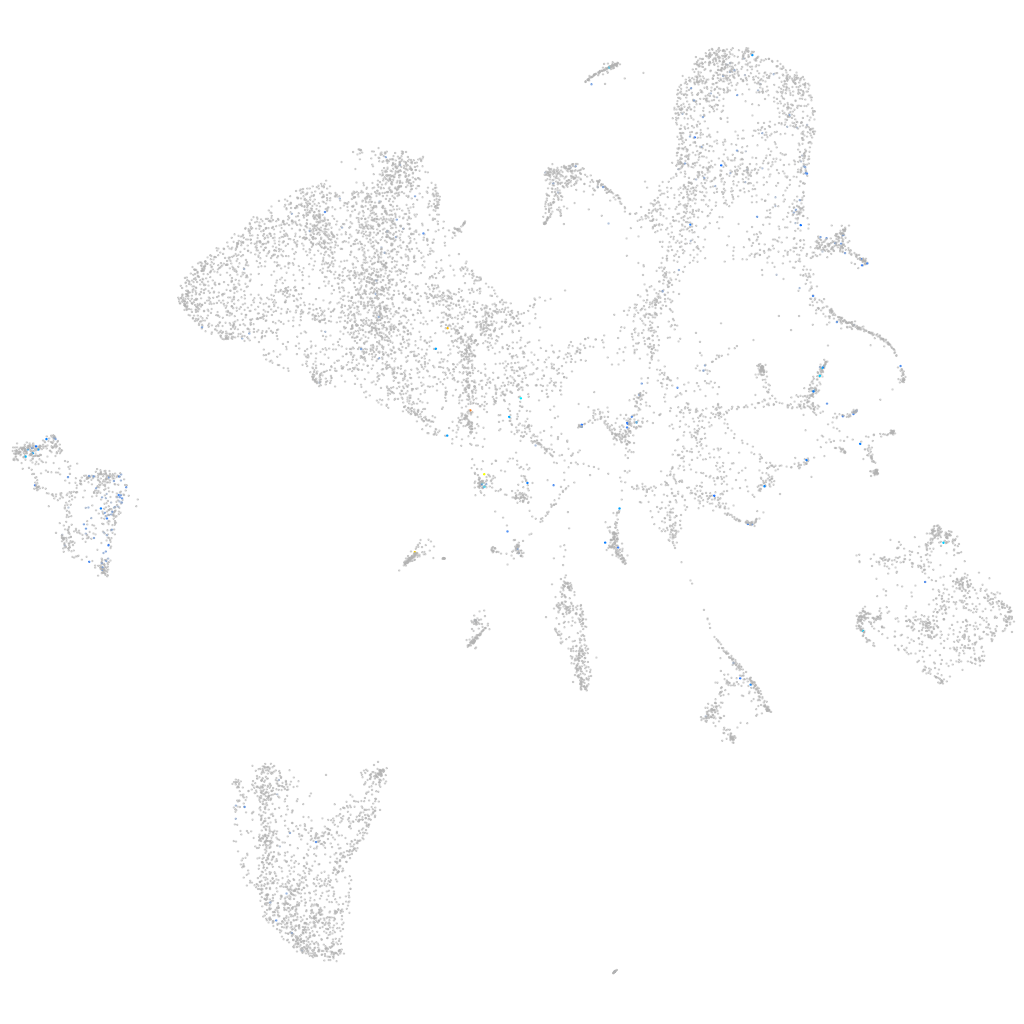
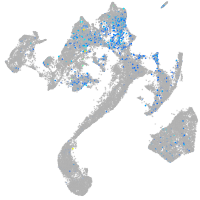
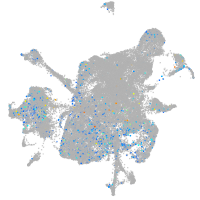
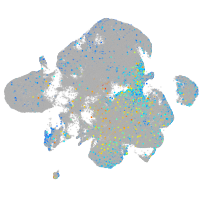
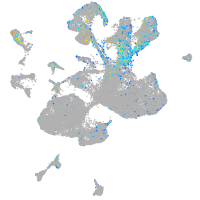
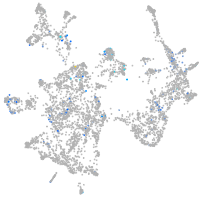
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tagapb | 0.292 | gamt | -0.068 |
| myo1f | 0.285 | gatm | -0.065 |
| irf8 | 0.268 | aqp12 | -0.064 |
| igfbp6b | 0.254 | bhmt | -0.063 |
| arhgef3l | 0.232 | gapdh | -0.060 |
| CU984600.2 | 0.208 | agxtb | -0.057 |
| RF00560 | 0.207 | mat1a | -0.057 |
| ccl34b.1 | 0.194 | vtnb | -0.057 |
| zgc:91999 | 0.189 | hao1 | -0.056 |
| AL831726.2 | 0.187 | serpinf2b | -0.054 |
| bcan | 0.180 | adka | -0.053 |
| casp8l2 | 0.180 | serpina1l | -0.052 |
| si:ch211-168d1.3 | 0.172 | zgc:112265 | -0.052 |
| gpr31 | 0.170 | cpn1 | -0.051 |
| si:dkey-27m7.4 | 0.170 | cfhl4 | -0.051 |
| kcnh2a | 0.165 | pygl | -0.051 |
| flrt1b | 0.145 | wu:fj16a03 | -0.051 |
| LOC101885859 | 0.144 | c9 | -0.051 |
| LOC103911631 | 0.142 | cfb | -0.050 |
| reep6 | 0.141 | grhprb | -0.050 |
| ano2 | 0.137 | ftcd | -0.050 |
| tlx1 | 0.135 | igfbp2a | -0.050 |
| LOC103912045 | 0.134 | tfa | -0.050 |
| CR361551.1 | 0.133 | vtna | -0.050 |
| BX510924.1 | 0.130 | LOC110437731 | -0.049 |
| itgb8 | 0.129 | hgd | -0.049 |
| si:ch73-190f9.4 | 0.127 | ttc36 | -0.049 |
| rin3 | 0.127 | ugp2a | -0.049 |
| LOC103911739 | 0.127 | fgg | -0.049 |
| BX005183.3 | 0.125 | itih2 | -0.049 |
| myh6 | 0.125 | shmt1 | -0.049 |
| wipf1b | 0.124 | serpinf2a | -0.049 |
| prickle2a | 0.123 | ppdpfa | -0.048 |
| zmp:0000000951 | 0.121 | zgc:123103 | -0.048 |
| tex33 | 0.121 | serpind1 | -0.048 |