kinesin family member 21A
ZFIN 
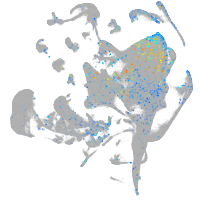
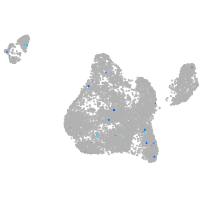
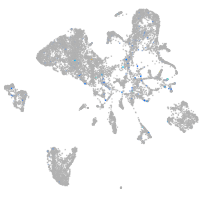
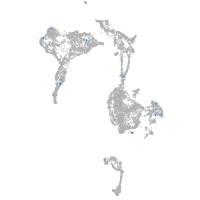
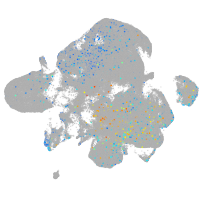
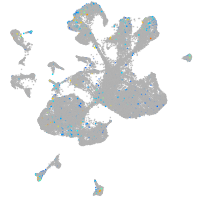
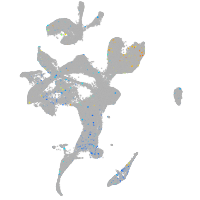
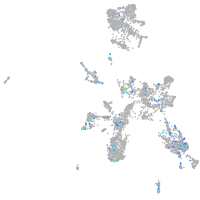
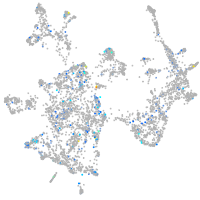
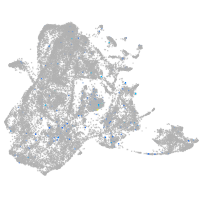
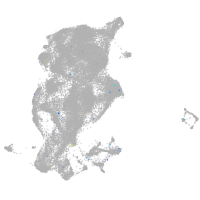
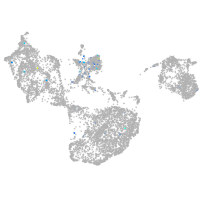
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tgm2a | 0.327 | hsp90ab1 | -0.236 |
| CABZ01072309.1 | 0.303 | pabpc1a | -0.213 |
| zgc:158296 | 0.289 | si:ch73-1a9.3 | -0.205 |
| myhz1.3 | 0.289 | hmgb2b | -0.204 |
| cavin4b | 0.283 | fthl27 | -0.202 |
| srl | 0.277 | ubc | -0.199 |
| tnnt3a | 0.273 | h3f3d | -0.196 |
| calm3a | 0.271 | si:ch211-222l21.1 | -0.195 |
| atp2a1 | 0.268 | hmgb2a | -0.195 |
| cav3 | 0.267 | ptmab | -0.193 |
| rtn2a | 0.267 | h2afvb | -0.192 |
| pygma | 0.264 | hnrnpaba | -0.189 |
| trdn | 0.263 | ran | -0.188 |
| fitm1 | 0.263 | khdrbs1a | -0.186 |
| acta1b | 0.262 | hmgn7 | -0.184 |
| casq2 | 0.261 | hmgn2 | -0.184 |
| aldoab | 0.260 | si:ch73-281n10.2 | -0.182 |
| si:ch211-156j16.1 | 0.259 | cirbpb | -0.181 |
| ckmb | 0.259 | actb2 | -0.179 |
| tmem38a | 0.259 | cirbpa | -0.177 |
| hhatla | 0.257 | hnrnpabb | -0.174 |
| XLOC-001975 | 0.257 | hmga1a | -0.173 |
| neb | 0.257 | anp32a | -0.172 |
| CABZ01072309.2 | 0.257 | syncrip | -0.172 |
| desma | 0.257 | tuba8l4 | -0.172 |
| ckma | 0.256 | cbx3a | -0.166 |
| ldb3b | 0.255 | anp32b | -0.165 |
| tpma | 0.254 | si:ch211-288g17.3 | -0.164 |
| klhl31 | 0.254 | hnrnpa0b | -0.164 |
| ryr1b | 0.254 | sumo3a | -0.163 |
| CABZ01078594.1 | 0.253 | seta | -0.161 |
| myom1a | 0.252 | h2afva | -0.160 |
| pgam2 | 0.252 | cfl1 | -0.159 |
| tmod4 | 0.252 | cx43.4 | -0.158 |
| gapdh | 0.251 | hdac1 | -0.155 |