potassium channel tetramerisation domain containing 12.2
ZFIN 
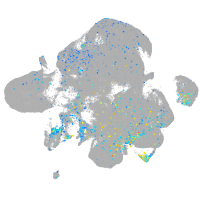
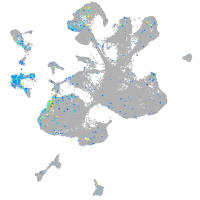
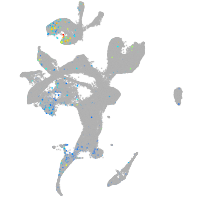
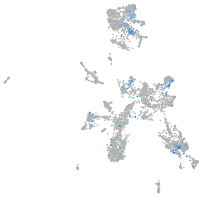
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tmem86a | 0.652 | ncl | -0.374 |
| adss | 0.628 | NC-002333.4 | -0.358 |
| zgc:153031 | 0.601 | banf1 | -0.346 |
| dusp28 | 0.593 | h1m | -0.346 |
| ppap2d | 0.577 | hnrnpa1b | -0.345 |
| dmtn | 0.568 | hnrnpaba | -0.338 |
| BX005254.1 | 0.559 | hmgb1a | -0.337 |
| mb | 0.557 | nasp | -0.323 |
| phactr3b | 0.551 | ppig | -0.319 |
| gria1b | 0.540 | gar1 | -0.317 |
| upf3a | 0.538 | ccna2 | -0.313 |
| zgc:153409 | 0.536 | cd2bp2 | -0.313 |
| myo18aa | 0.534 | srrt | -0.302 |
| igsf8 | 0.534 | sde2 | -0.300 |
| cnrip1a | 0.531 | chaf1a | -0.299 |
| pank2 | 0.522 | slu7 | -0.295 |
| emd | 0.521 | dlgap5 | -0.292 |
| herc1 | 0.516 | knop1 | -0.291 |
| cyp4v7 | 0.512 | akap12b | -0.291 |
| elf1 | 0.512 | hnrnpub | -0.290 |
| si:dkeyp-117h8.2 | 0.507 | snrpa | -0.289 |
| stambpa | 0.504 | ebna1bp2 | -0.287 |
| proca1 | 0.503 | hnrnpd | -0.286 |
| gsnb | 0.501 | smarca4a | -0.286 |
| per3 | 0.501 | rps18 | -0.283 |
| stat1a | 0.498 | tbx16 | -0.283 |
| trim55b | 0.493 | AL935174.5 | -0.281 |
| LOC108190923 | 0.493 | cdk11b | -0.281 |
| kalrnb | 0.493 | mphosph10 | -0.277 |
| gpr176 | 0.493 | apoeb | -0.270 |
| CABZ01030107.1 | 0.490 | pcna | -0.267 |
| si:ch73-177h5.2 | 0.488 | marcksl1b | -0.265 |
| RASSF5 | 0.484 | marcksb | -0.265 |
| zgc:193726 | 0.484 | brd3a | -0.264 |
| dnajc18 | 0.483 | nop58 | -0.263 |