"potassium channel, subfamily K, member 18"
ZFIN 
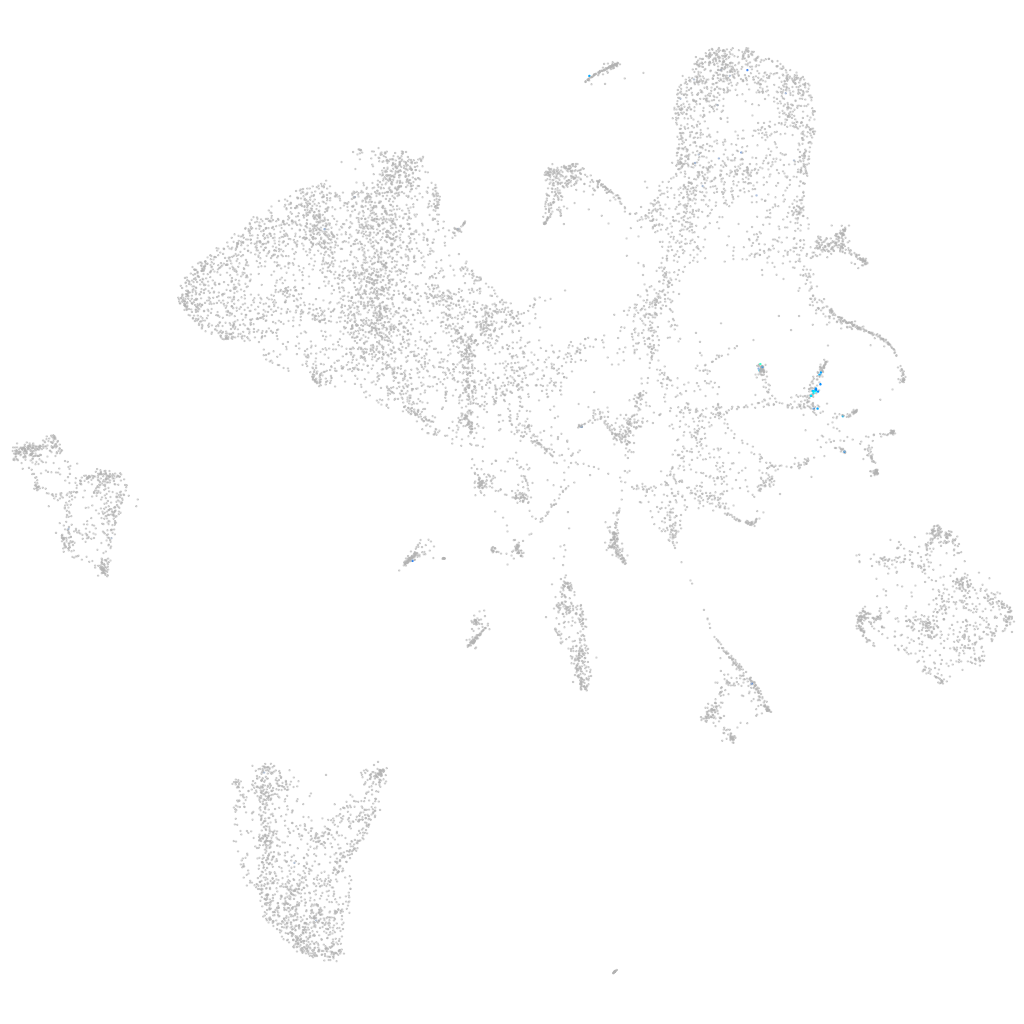
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 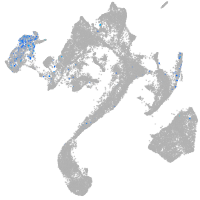 |
mural cells / non-skeletal muscle 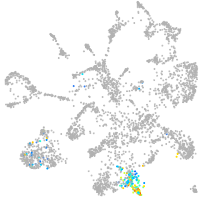 |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural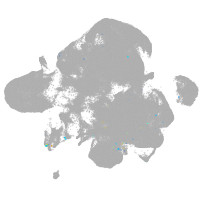 |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line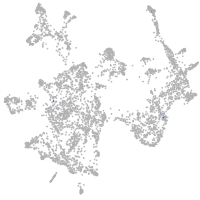 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pyyb | 0.347 | ahcy | -0.055 |
| plcxd3 | 0.237 | fabp3 | -0.051 |
| npy2rl | 0.224 | gamt | -0.046 |
| gng13b | 0.218 | aldh7a1 | -0.043 |
| xpnpep2 | 0.214 | rps18 | -0.042 |
| ccka | 0.213 | bhmt | -0.042 |
| AL935199.1 | 0.210 | rplp2l | -0.041 |
| si:dkey-59l11.10 | 0.206 | rps20 | -0.041 |
| rem2 | 0.202 | hdlbpa | -0.041 |
| LOC110439478 | 0.202 | eif4ebp1 | -0.041 |
| scgn | 0.199 | rpl6 | -0.040 |
| c2cd4a | 0.199 | si:dkey-16p21.8 | -0.040 |
| rprmb | 0.198 | gatm | -0.039 |
| tmem178b | 0.197 | hsdl2 | -0.039 |
| LOC103910220 | 0.196 | mat1a | -0.038 |
| si:ch73-160i9.2 | 0.195 | gapdh | -0.038 |
| egr4 | 0.194 | rpl4 | -0.038 |
| arxa | 0.191 | rps12 | -0.037 |
| lrrc38b | 0.190 | hmgb3b | -0.037 |
| CABZ01001434.1 | 0.190 | cebpd | -0.036 |
| adora2aa | 0.185 | shmt1 | -0.036 |
| ptger3 | 0.185 | apoc1 | -0.036 |
| helz2 | 0.183 | gnmt | -0.036 |
| abcc8 | 0.181 | hspd1 | -0.036 |
| anxa5a | 0.176 | rpl5a | -0.035 |
| ms4a17a.10 | 0.174 | tktb | -0.035 |
| ldb3a | 0.173 | grhprb | -0.035 |
| TCIM (1 of many) | 0.172 | rpl22l1 | -0.035 |
| rasd4 | 0.170 | rgrb | -0.035 |
| fev | 0.169 | cx32.3 | -0.034 |
| b4galt2 | 0.169 | slc25a34 | -0.034 |
| b4galnt2.2 | 0.168 | tfa | -0.034 |
| tango2 | 0.164 | agxta | -0.034 |
| LOC110440005 | 0.163 | si:dkey-151g10.6 | -0.034 |
| BX323596.2 | 0.162 | wu:fj16a03 | -0.034 |