jumonji domain containing 4
ZFIN 
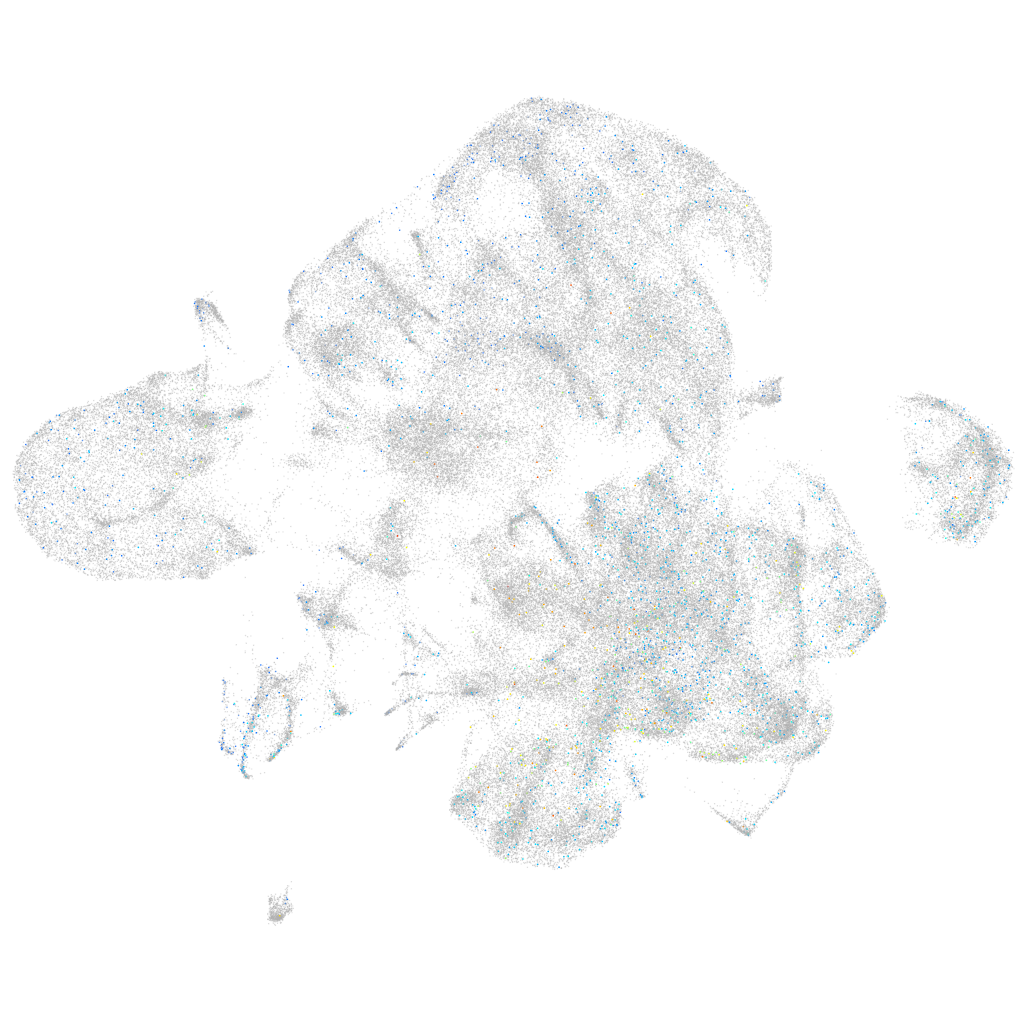
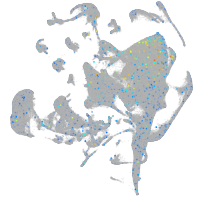
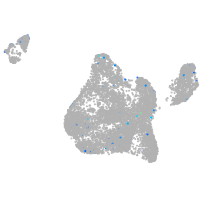
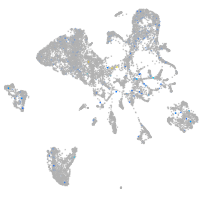
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gng3 | 0.067 | hmgb2a | -0.051 |
| ywhag2 | 0.062 | id1 | -0.039 |
| atp6v1e1b | 0.060 | hmga1a | -0.039 |
| zgc:65894 | 0.060 | msna | -0.037 |
| stmn2a | 0.059 | mdka | -0.037 |
| snap25a | 0.059 | pcna | -0.037 |
| sncb | 0.059 | stmn1a | -0.037 |
| stmn1b | 0.058 | rplp2l | -0.036 |
| stxbp1a | 0.058 | rplp1 | -0.035 |
| calm1a | 0.057 | mki67 | -0.034 |
| mllt11 | 0.056 | XLOC-003690 | -0.034 |
| atp6v1g1 | 0.055 | her15.1 | -0.034 |
| tuba1c | 0.055 | sox3 | -0.034 |
| rnasekb | 0.054 | si:dkey-151g10.6 | -0.034 |
| calm2a | 0.054 | tuba8l | -0.033 |
| gapdhs | 0.053 | XLOC-003689 | -0.033 |
| vamp2 | 0.052 | rps12 | -0.033 |
| elavl4 | 0.052 | ccnd1 | -0.032 |
| map1aa | 0.052 | si:ch73-281n10.2 | -0.032 |
| ckbb | 0.052 | ccna2 | -0.031 |
| atp6v0cb | 0.051 | chaf1a | -0.031 |
| fez1 | 0.051 | dut | -0.031 |
| tpi1b | 0.050 | anp32b | -0.031 |
| tmsb2 | 0.050 | selenoh | -0.030 |
| gng2 | 0.050 | COX7A2 (1 of many) | -0.030 |
| gap43 | 0.050 | hmgb2b | -0.030 |
| gpm6ab | 0.049 | sox19a | -0.030 |
| calm3a | 0.049 | XLOC-003692 | -0.030 |
| rtn1b | 0.049 | sdc4 | -0.030 |
| eno2 | 0.049 | npm1a | -0.030 |
| ywhah | 0.049 | notch3 | -0.030 |
| rtn1a | 0.049 | mad2l1 | -0.030 |
| h2afx1 | 0.049 | rps20 | -0.029 |
| si:dkey-276j7.1 | 0.048 | ranbp1 | -0.029 |
| hist2h2l | 0.048 | vamp3 | -0.029 |