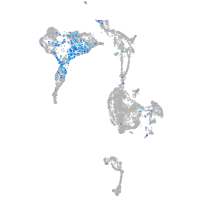
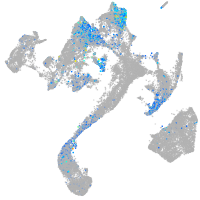
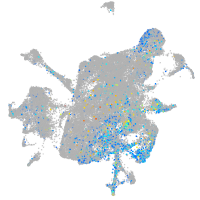
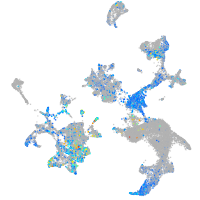
integrin alpha 4
ZFIN 
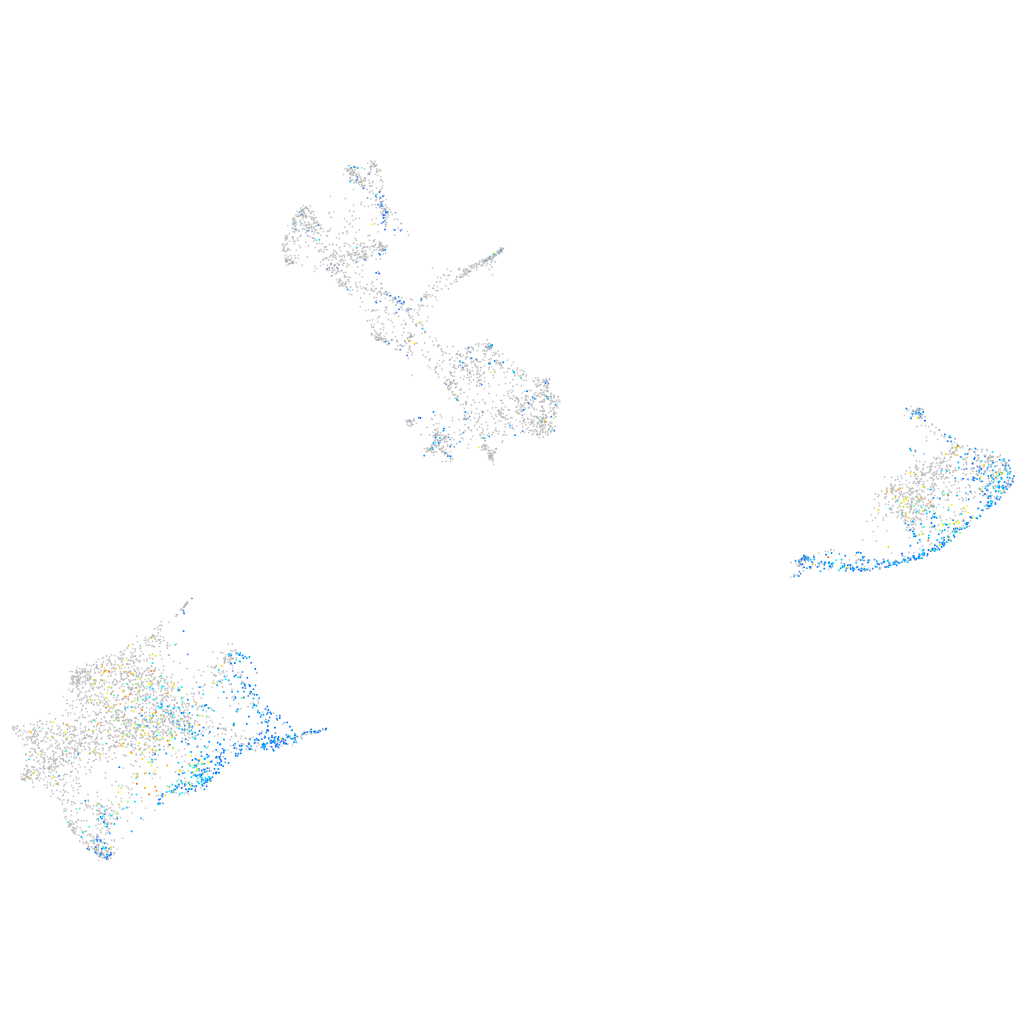
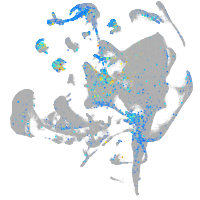
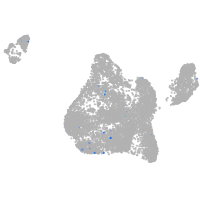
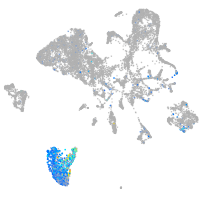
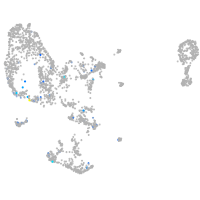
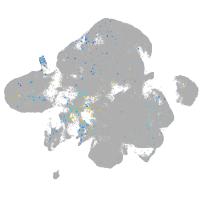
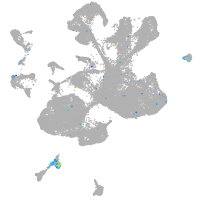
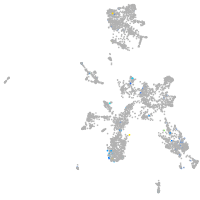
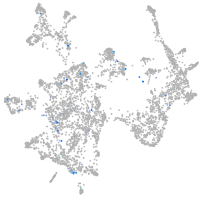
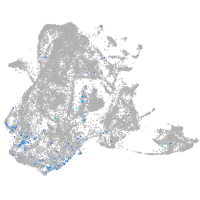
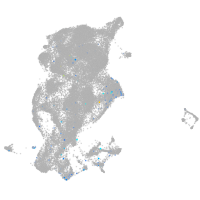
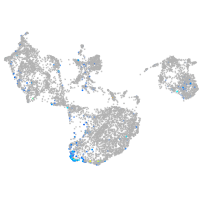
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slc2a11b | 0.266 | defbl1 | -0.166 |
| pah | 0.262 | apoda.1 | -0.162 |
| tyr | 0.252 | mdkb | -0.155 |
| mitfa | 0.252 | gpnmb | -0.148 |
| qdpra | 0.248 | lypc | -0.147 |
| canx | 0.245 | si:ch211-256m1.8 | -0.142 |
| SPAG9 | 0.237 | si:dkey-197i20.6 | -0.141 |
| opn5 | 0.233 | akap12a | -0.130 |
| tmem243a | 0.229 | si:ch73-89b15.3 | -0.128 |
| pcdh10a | 0.227 | ptmaa | -0.125 |
| tspan36 | 0.226 | ponzr1 | -0.124 |
| LOC103910009 | 0.225 | s100v2 | -0.124 |
| pcdh9 | 0.222 | alx4a | -0.119 |
| phlda1 | 0.220 | unm-sa821 | -0.117 |
| atp11a | 0.215 | sytl2b | -0.116 |
| pim1 | 0.214 | alx1 | -0.116 |
| tspan10 | 0.213 | guk1a | -0.114 |
| syngr1a | 0.213 | mdka | -0.113 |
| lamp1a | 0.213 | alx4b | -0.112 |
| rab34b | 0.207 | si:ch73-1a9.3 | -0.111 |
| si:zfos-943e10.1 | 0.205 | pvalb1 | -0.109 |
| sprb | 0.200 | hmgb1b | -0.108 |
| rab38 | 0.199 | slc25a36a | -0.108 |
| bace2 | 0.198 | cx43 | -0.107 |
| sypl2b | 0.197 | cdh11 | -0.106 |
| naga | 0.196 | ppdpfb | -0.105 |
| mocos | 0.196 | prx | -0.104 |
| atp6v0ca | 0.194 | gpm6aa | -0.103 |
| slc24a4a | 0.193 | CABZ01072077.1 | -0.103 |
| cdh1 | 0.193 | CT737162.3 | -0.100 |
| myo5aa | 0.193 | sdc2 | -0.099 |
| rabl6b | 0.192 | tpd52l1 | -0.099 |
| tdh | 0.192 | stmn1b | -0.099 |
| kcnj13 | 0.192 | nova2 | -0.099 |
| rgs2 | 0.192 | tuba1b | -0.097 |