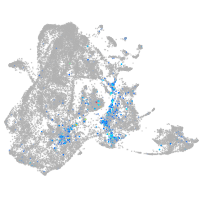
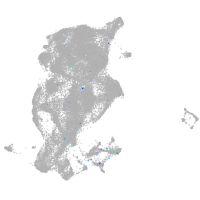
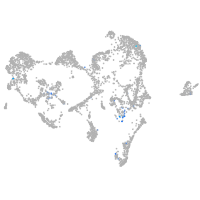
ISL LIM homeobox 2b
ZFIN 
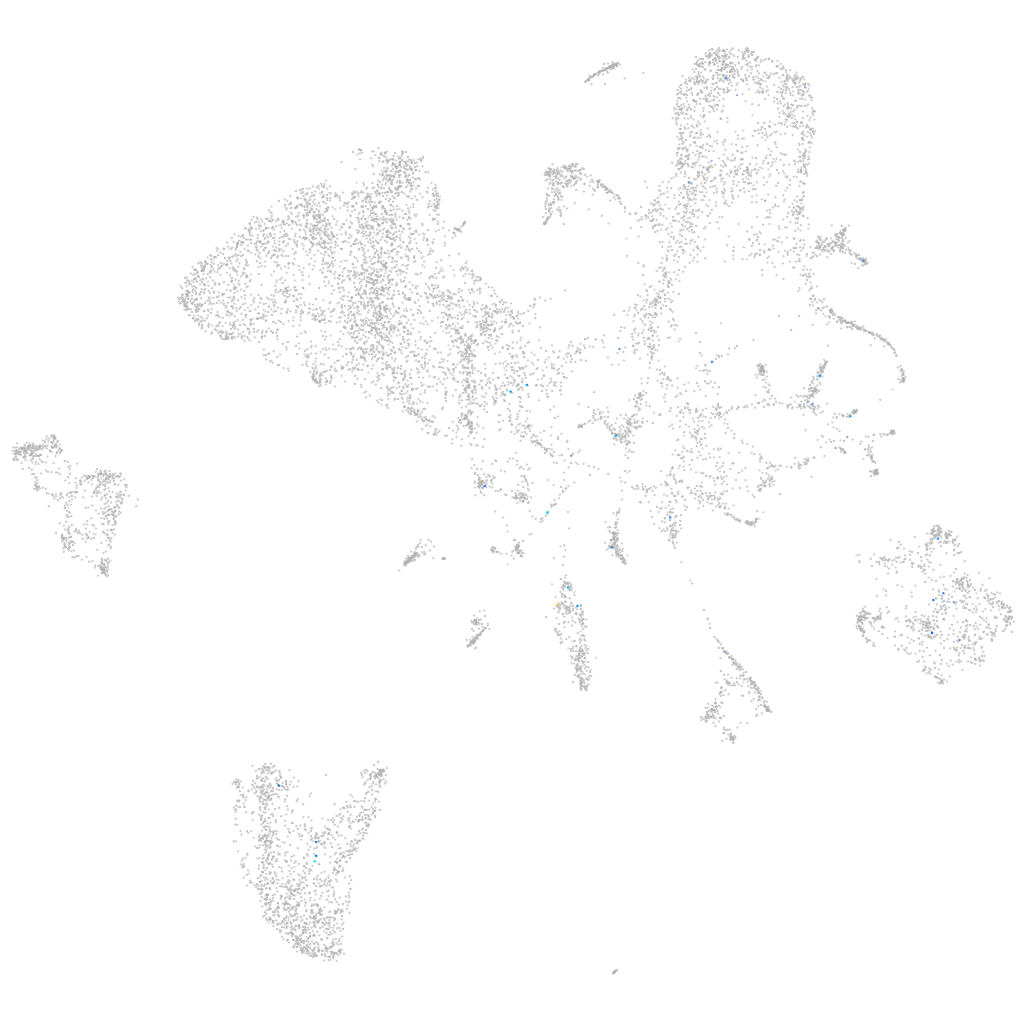
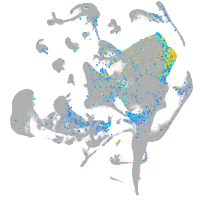
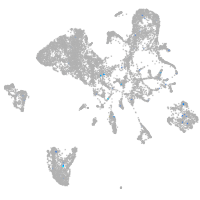
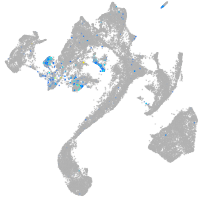
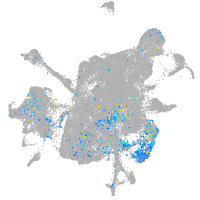
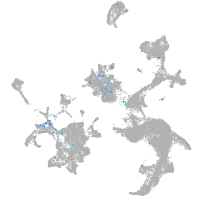
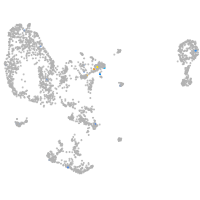
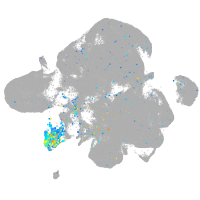
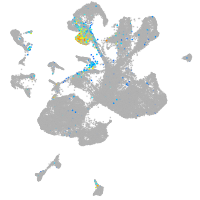
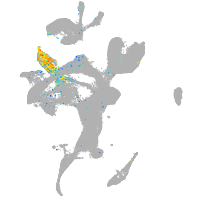
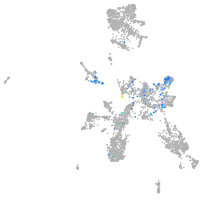
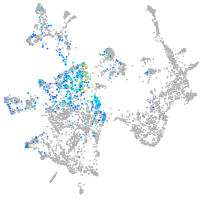
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC110440218 | 0.339 | ckba | -0.038 |
| krt98 | 0.329 | atp5pb | -0.036 |
| XLOC-022188 | 0.299 | agxta | -0.034 |
| kif26bb | 0.286 | mgst1.2 | -0.034 |
| LOC110438054 | 0.276 | scp2a | -0.034 |
| tmem151bb | 0.247 | gstr | -0.034 |
| arxb | 0.231 | sdr16c5b | -0.033 |
| sypl1 | 0.223 | cyb5a | -0.033 |
| kcnk10b | 0.198 | mdh1aa | -0.032 |
| wdr47b | 0.194 | aldh6a1 | -0.032 |
| asic1c | 0.189 | mt-nd1 | -0.032 |
| chata | 0.184 | atp5mc3b | -0.032 |
| ksr1a | 0.182 | gsta.1 | -0.032 |
| LOC103911631 | 0.181 | eno3 | -0.032 |
| CR854980.1 | 0.176 | sod1 | -0.032 |
| LOC108183341 | 0.172 | acaa2 | -0.031 |
| vsig10l2 | 0.170 | abat | -0.031 |
| BX547930.4 | 0.169 | haao | -0.031 |
| CABZ01072309.2 | 0.168 | ndufs4 | -0.031 |
| LOC100000890 | 0.168 | ttc36 | -0.031 |
| prph | 0.167 | fabp10a | -0.031 |
| CU861477.1 | 0.165 | pgm1 | -0.031 |
| atp6v0a2a | 0.164 | hpda | -0.031 |
| si:dkey-110g7.8 | 0.160 | phyhd1 | -0.031 |
| eno2 | 0.154 | tdo2a | -0.031 |
| BX927257.1 | 0.151 | chchd10 | -0.030 |
| or137-9 | 0.151 | uox | -0.030 |
| BX324132.2 | 0.151 | ttr | -0.030 |
| gdap1l1 | 0.149 | rgrb | -0.030 |
| LOC110437921 | 0.147 | dldh | -0.030 |
| stx12l | 0.147 | cat | -0.030 |
| insyn1 | 0.145 | rmdn1 | -0.030 |
| mtmr7b | 0.145 | decr1 | -0.030 |
| znf998 | 0.144 | cbr1l | -0.030 |
| LOC108183423 | 0.143 | prdx3 | -0.030 |