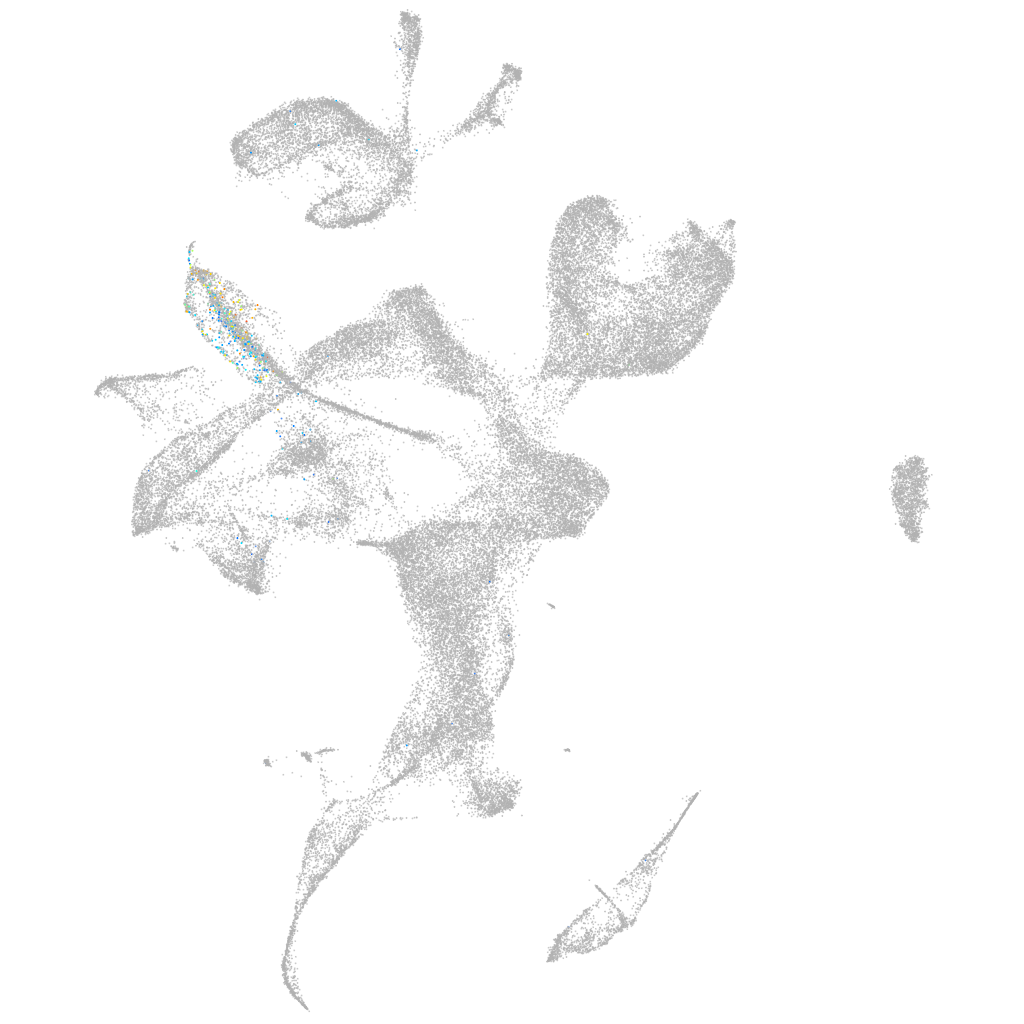
iroquois homeobox 4b
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory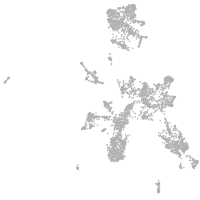 |
otic / lateral line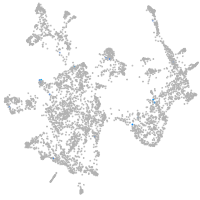 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells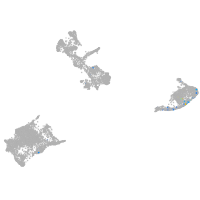 |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| irx4a | 0.350 | hmgb2a | -0.081 |
| rbpms2a | 0.281 | fabp7a | -0.048 |
| isl2b | 0.242 | mdka | -0.046 |
| islr2 | 0.232 | anp32e | -0.045 |
| ebf3a | 0.226 | msi1 | -0.042 |
| rbpms2b | 0.225 | hmga1a | -0.041 |
| inab | 0.223 | ahcy | -0.041 |
| irx2a | 0.212 | pcna | -0.041 |
| pou4f1 | 0.208 | neurod4 | -0.040 |
| pou6f2 | 0.206 | si:ch73-281n10.2 | -0.040 |
| zgc:158291 | 0.200 | dut | -0.039 |
| xpr1a | 0.196 | crx | -0.039 |
| gfi1ab | 0.194 | selenoh | -0.038 |
| BX247868.1 | 0.186 | rrm1 | -0.038 |
| grin2ab | 0.178 | mki67 | -0.038 |
| gap43 | 0.174 | stmn1a | -0.038 |
| id4 | 0.173 | otx5 | -0.038 |
| shisal1a | 0.163 | lbr | -0.038 |
| stmn2a | 0.159 | ccnd1 | -0.038 |
| nrn1a | 0.158 | nutf2l | -0.038 |
| rtn1b | 0.157 | syt5b | -0.038 |
| tmsb | 0.157 | msna | -0.038 |
| cplx2l | 0.155 | chaf1a | -0.037 |
| rab6ba | 0.153 | tuba8l | -0.037 |
| stmn2b | 0.152 | ccna2 | -0.037 |
| stmn4l | 0.149 | dek | -0.037 |
| frem2a | 0.147 | her15.1 | -0.035 |
| pou4f2 | 0.144 | eef1da | -0.035 |
| ppp1r14ba | 0.144 | mcm7 | -0.035 |
| stmn4 | 0.144 | ddah2 | -0.034 |
| gpr37b | 0.142 | COX7A2 (1 of many) | -0.034 |
| GNB4 | 0.141 | cks1b | -0.034 |
| eno2 | 0.138 | lig1 | -0.034 |
| oaz2b | 0.138 | rpa3 | -0.033 |
| cacna1aa | 0.135 | rrm2 | -0.033 |