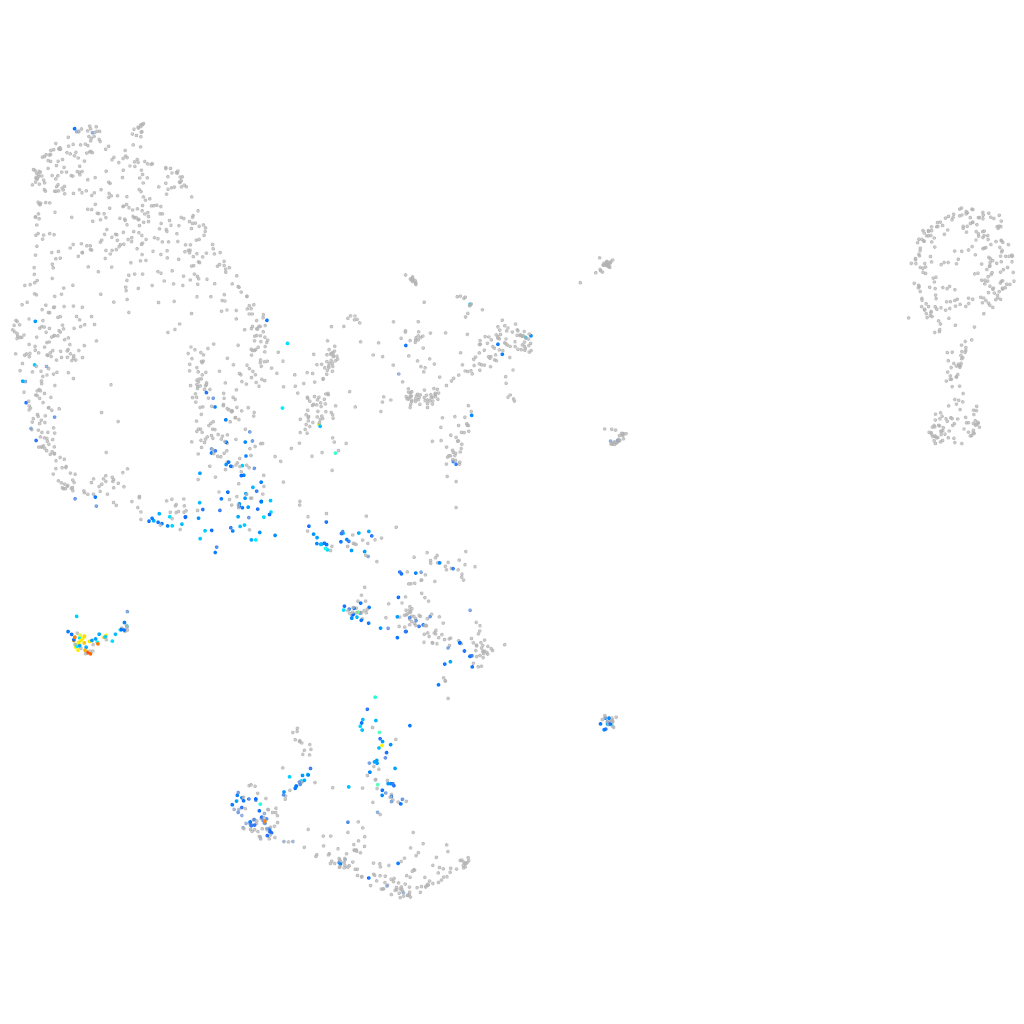
IQ motif containing G
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| capsla | 0.582 | gapdh | -0.214 |
| enkur | 0.576 | alpl | -0.200 |
| si:dkey-148f10.4 | 0.560 | dpys | -0.197 |
| zgc:195356 | 0.546 | si:dkey-33i11.9 | -0.188 |
| zgc:55461 | 0.528 | epdl2 | -0.187 |
| si:ch211-155m12.5 | 0.524 | agxtb | -0.187 |
| tbata | 0.521 | gpt2l | -0.184 |
| pacrg | 0.517 | phyhd1 | -0.183 |
| LOC100329490 | 0.511 | cx32.3 | -0.180 |
| si:dkey-83m22.7 | 0.511 | gcshb | -0.178 |
| spata18 | 0.506 | slc16a1b | -0.174 |
| LOC103911624 | 0.504 | pnp6 | -0.173 |
| si:dkeyp-46h3.1 | 0.499 | hao1 | -0.169 |
| spata4 | 0.495 | ASS1 | -0.169 |
| smkr1 | 0.493 | aco1 | -0.168 |
| cdc25d | 0.492 | nipsnap3a | -0.168 |
| si:ch211-163l21.7 | 0.488 | lgmn | -0.167 |
| pih1d3 | 0.477 | ctsla | -0.167 |
| mycbp | 0.475 | cubn | -0.165 |
| cfap45 | 0.473 | rps18 | -0.165 |
| tekt2 | 0.473 | slc5a12 | -0.165 |
| tekt4 | 0.471 | pck2 | -0.165 |
| dnali1 | 0.470 | haao | -0.163 |
| CFAP77 | 0.469 | slc26a6l | -0.162 |
| ccdc114 | 0.465 | g6pca.2 | -0.162 |
| capslb | 0.462 | si:ch211-201h21.5 | -0.159 |
| si:ch73-81k8.2 | 0.457 | rgn | -0.159 |
| ccdc169 | 0.457 | slc34a1a | -0.159 |
| si:dkey-11c5.11 | 0.457 | gstt1a | -0.159 |
| erich3 | 0.453 | aldob | -0.158 |
| spag6 | 0.452 | dhrs9 | -0.156 |
| ropn1l | 0.448 | slc5a11 | -0.156 |
| RSPH1 | 0.448 | scpep1 | -0.154 |
| meig1 | 0.447 | fbp1b | -0.154 |
| si:dkey-42p14.3 | 0.445 | adh8b | -0.154 |