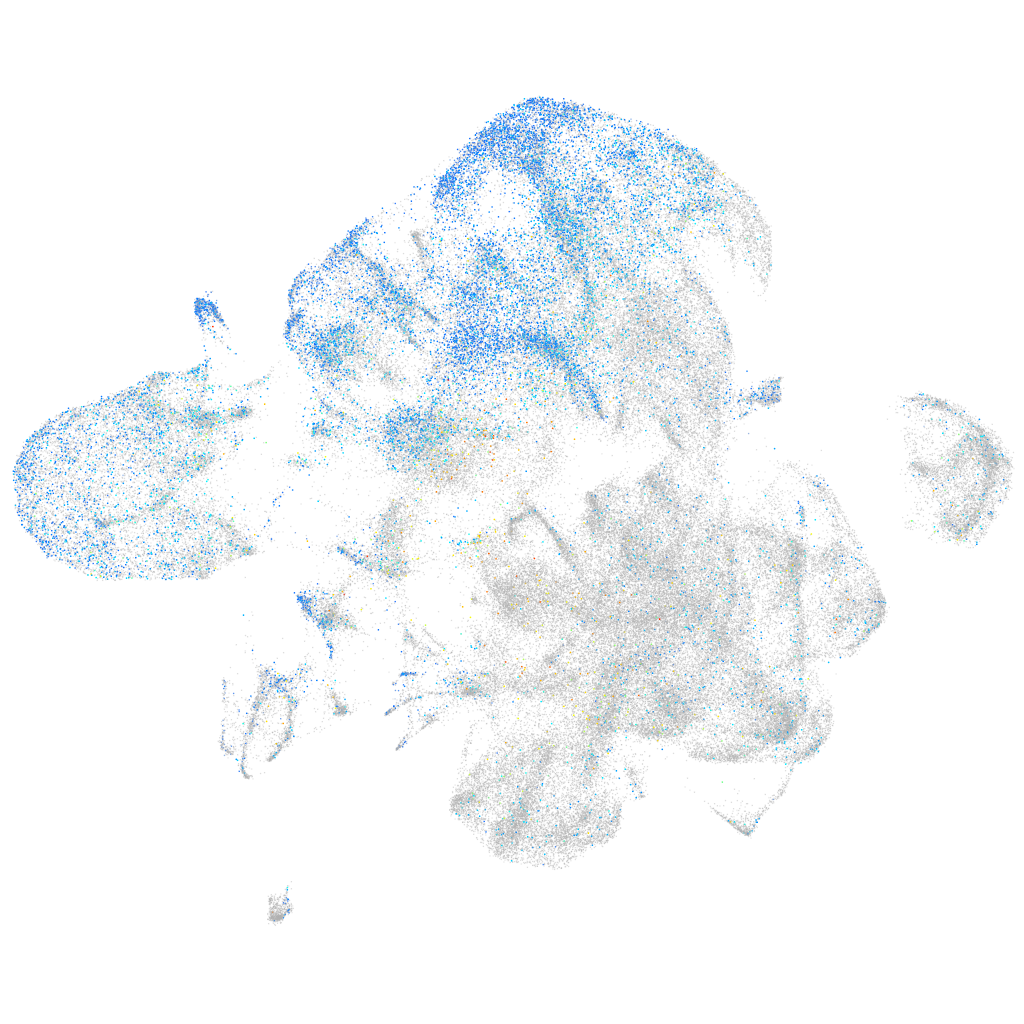
importin 9
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pcna | 0.242 | elavl3 | -0.170 |
| dut | 0.216 | rtn1a | -0.170 |
| rrm1 | 0.213 | stmn1b | -0.164 |
| rpa3 | 0.203 | gpm6aa | -0.145 |
| mcm7 | 0.200 | gpm6ab | -0.143 |
| chaf1a | 0.199 | ckbb | -0.142 |
| fen1 | 0.197 | gng3 | -0.140 |
| anp32b | 0.196 | sncb | -0.132 |
| nutf2l | 0.195 | atp6v0cb | -0.130 |
| hmgb2a | 0.194 | rtn1b | -0.130 |
| rpa2 | 0.194 | tmsb | -0.129 |
| tuba8l4 | 0.193 | rnasekb | -0.125 |
| nasp | 0.191 | vamp2 | -0.122 |
| ranbp1 | 0.191 | elavl4 | -0.122 |
| stmn1a | 0.191 | si:dkeyp-75h12.5 | -0.122 |
| rrm2 | 0.190 | zc4h2 | -0.121 |
| id1 | 0.183 | ptmaa | -0.118 |
| banf1 | 0.182 | h2afx1 | -0.117 |
| selenoh | 0.182 | tubb5 | -0.117 |
| lig1 | 0.179 | tuba1c | -0.117 |
| ccna2 | 0.179 | fez1 | -0.116 |
| dhfr | 0.178 | ppdpfb | -0.114 |
| zgc:110540 | 0.178 | gng2 | -0.114 |
| ccnd1 | 0.176 | ywhag2 | -0.114 |
| pa2g4b | 0.175 | stx1b | -0.114 |
| CABZ01005379.1 | 0.175 | stmn2a | -0.113 |
| lbr | 0.175 | calm1a | -0.112 |
| slbp | 0.173 | dpysl3 | -0.111 |
| dek | 0.173 | mllt11 | -0.110 |
| mibp | 0.173 | ywhah | -0.110 |
| ssrp1a | 0.172 | atp6v1e1b | -0.109 |
| mcm4 | 0.171 | stxbp1a | -0.108 |
| snu13b | 0.170 | myt1b | -0.108 |
| tuba8l | 0.169 | atp6v1g1 | -0.108 |
| rnaseh2a | 0.168 | kdm6bb | -0.108 |