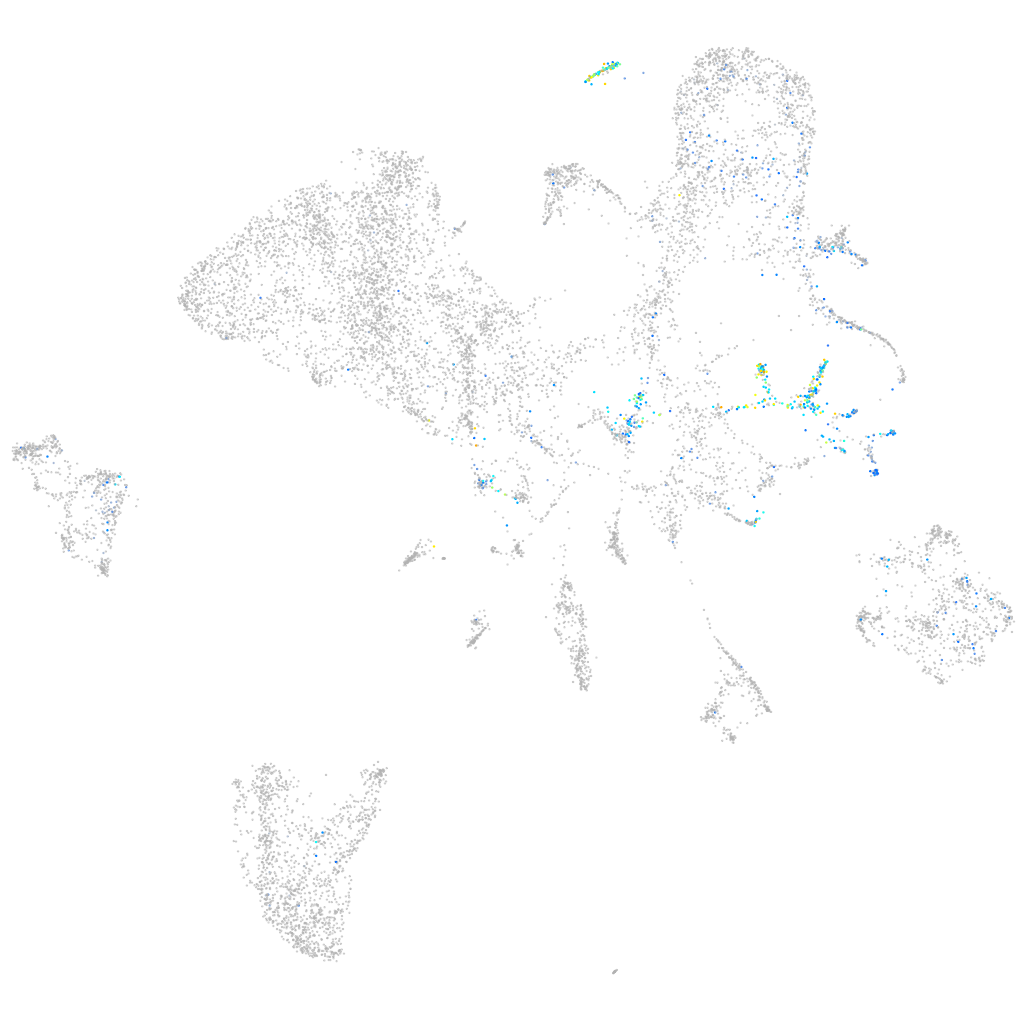
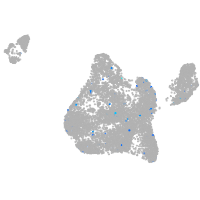
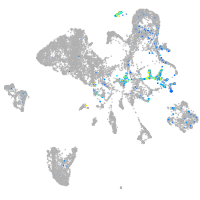
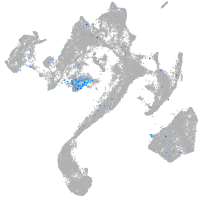
insulinoma-associated 1b
ZFIN 
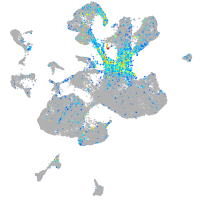
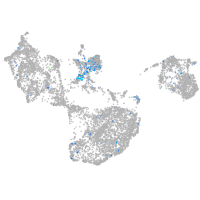
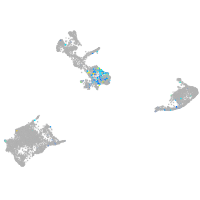
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.620 | gamt | -0.204 |
| neurod1 | 0.551 | ahcy | -0.190 |
| zgc:101731 | 0.513 | gapdh | -0.190 |
| egr4 | 0.509 | fabp3 | -0.187 |
| dll4 | 0.503 | bhmt | -0.179 |
| mir7a-1 | 0.498 | gatm | -0.178 |
| fev | 0.489 | hdlbpa | -0.173 |
| pcsk1nl | 0.466 | mat1a | -0.173 |
| nkx2.2a | 0.453 | nupr1b | -0.163 |
| id4 | 0.451 | aldob | -0.163 |
| mir375-2 | 0.450 | aqp12 | -0.161 |
| ret | 0.449 | fbp1b | -0.161 |
| insm1a | 0.449 | apoa1b | -0.159 |
| hepacam2 | 0.447 | aldh6a1 | -0.157 |
| c2cd4a | 0.445 | aldh7a1 | -0.157 |
| capsla | 0.436 | cx32.3 | -0.157 |
| si:ch73-160i9.2 | 0.432 | apoa4b.1 | -0.157 |
| pax4 | 0.411 | agxtb | -0.154 |
| adcyap1a | 0.409 | apoc2 | -0.152 |
| TCIM (1 of many) | 0.408 | gnmt | -0.152 |
| elovl1a | 0.406 | apoc1 | -0.151 |
| lysmd2 | 0.398 | mgst1.2 | -0.148 |
| rgs16 | 0.396 | si:dkey-16p21.8 | -0.148 |
| tmsb | 0.396 | tfa | -0.147 |
| pax6b | 0.389 | agxta | -0.147 |
| vat1 | 0.388 | afp4 | -0.147 |
| syt1a | 0.384 | wu:fj16a03 | -0.146 |
| trpa1b | 0.383 | apoa2 | -0.145 |
| ccka | 0.378 | gstr | -0.142 |
| penka | 0.375 | serpina1 | -0.140 |
| calm1b | 0.374 | ttc36 | -0.139 |
| zgc:165481 | 0.373 | apobb.1 | -0.139 |
| dusp2 | 0.372 | apom | -0.138 |
| dlb | 0.370 | ftcd | -0.138 |
| pcsk1 | 0.366 | serpina1l | -0.138 |