insulinoma-associated 1a
ZFIN 
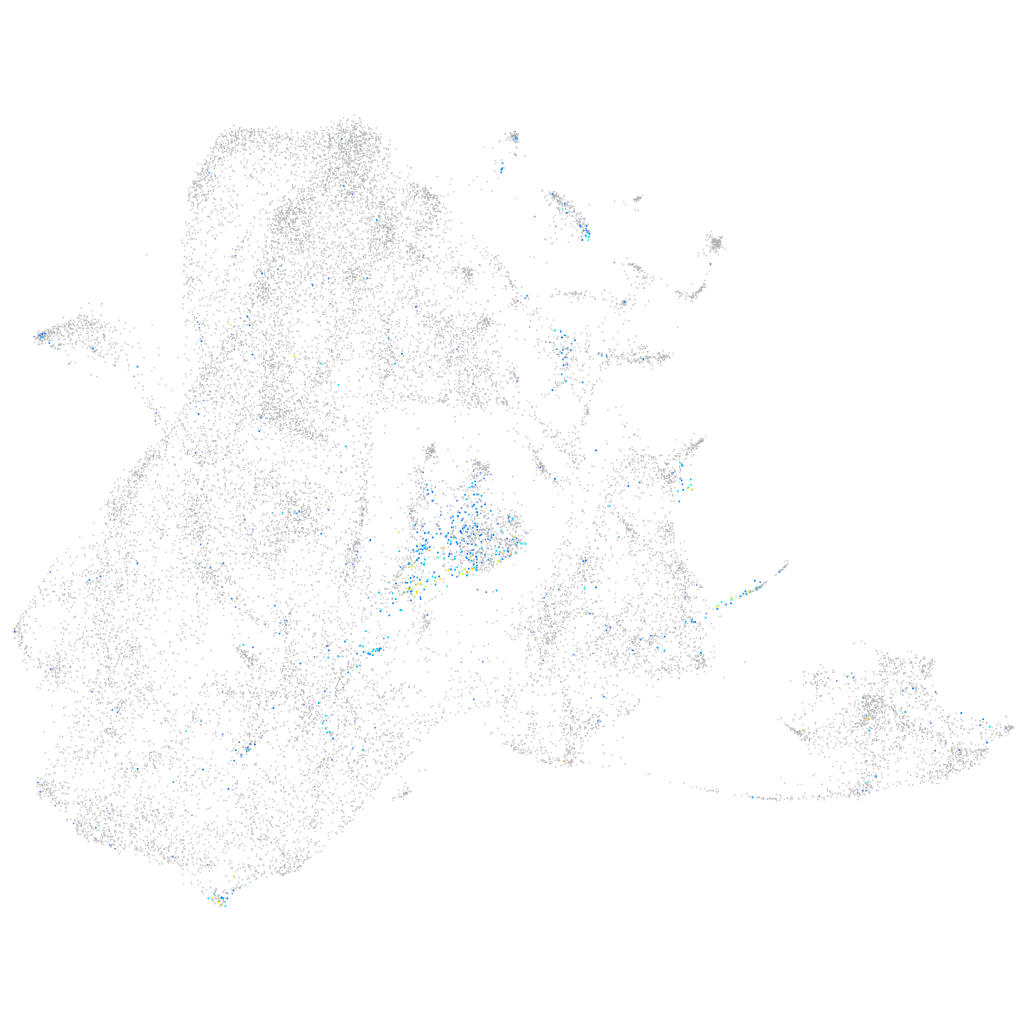
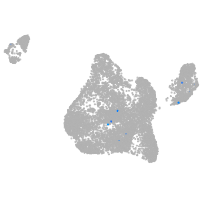
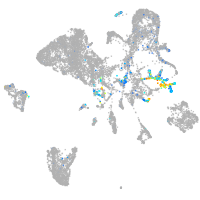
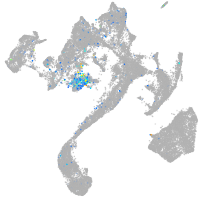
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dlb | 0.411 | cfl1l | -0.134 |
| insm1b | 0.354 | pfn1 | -0.106 |
| srrm4 | 0.319 | tmsb1 | -0.098 |
| myt1b | 0.319 | krt4 | -0.094 |
| elavl3 | 0.302 | cldn1 | -0.090 |
| gadd45gb.1 | 0.297 | cyt1 | -0.088 |
| myt1a | 0.281 | s100a10b | -0.080 |
| rtn1a | 0.278 | cebpd | -0.078 |
| ebf2 | 0.269 | myh9a | -0.077 |
| si:dkey-56m19.5 | 0.266 | lgals3b | -0.076 |
| neurod4 | 0.265 | ecrg4b | -0.076 |
| dla | 0.263 | arhgef5 | -0.075 |
| tuba1a | 0.257 | zgc:162730 | -0.074 |
| hes6 | 0.257 | csrp1a | -0.074 |
| nova2 | 0.257 | actb2 | -0.072 |
| ascl1a | 0.252 | hdlbpa | -0.072 |
| stmn1b | 0.247 | tp63 | -0.071 |
| tmsb | 0.241 | cebpb | -0.071 |
| tp53inp2 | 0.241 | pleca | -0.071 |
| LOC798783 | 0.238 | si:ch211-166a6.5 | -0.070 |
| inavaa | 0.233 | anxa1a | -0.070 |
| tuba1c | 0.231 | si:dkey-16p21.8 | -0.070 |
| CU467822.1 | 0.218 | zgc:101810 | -0.069 |
| her4.2 | 0.215 | pak2a | -0.069 |
| scrt2 | 0.215 | arpc2 | -0.068 |
| BX530077.1 | 0.215 | col11a1a | -0.067 |
| nhlh2 | 0.214 | tmem238a | -0.067 |
| neurod1 | 0.213 | tmsb4x | -0.066 |
| gpm6aa | 0.212 | dnase1l4.1 | -0.066 |
| CU634008.1 | 0.212 | zgc:153867 | -0.066 |
| epb41a | 0.208 | cldni | -0.066 |
| abhd6a | 0.207 | sparc | -0.065 |
| si:ch211-137a8.4 | 0.206 | capns1a | -0.064 |
| sox1b | 0.205 | tagln2 | -0.064 |
| vamp2 | 0.203 | krt8 | -0.064 |