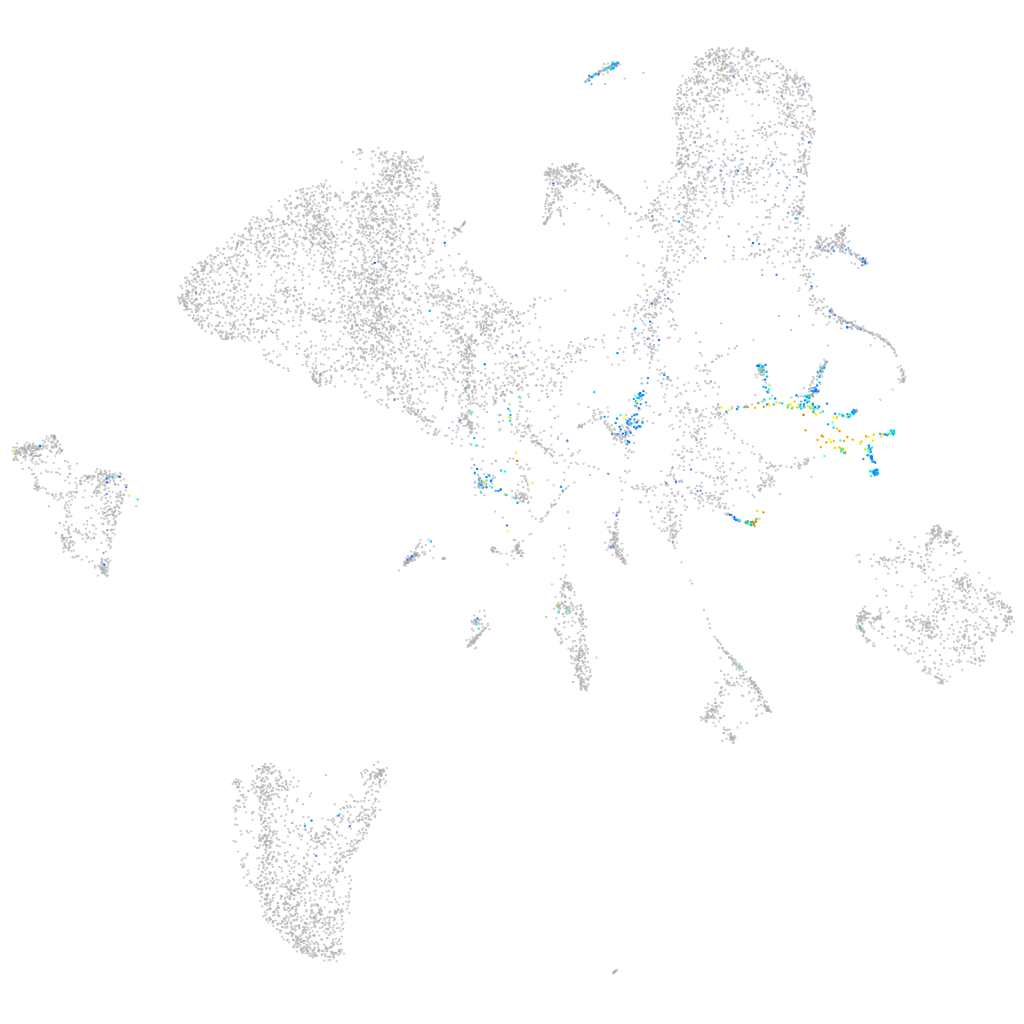
insulinoma-associated 1a
ZFIN 
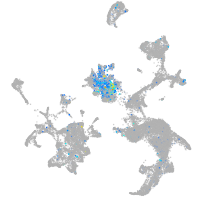
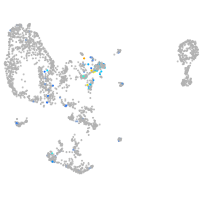
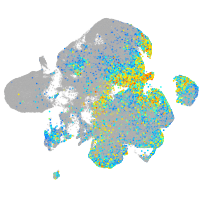
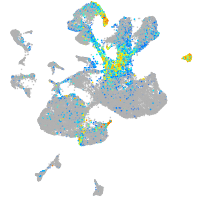
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| neurod1 | 0.642 | aldob | -0.216 |
| scg3 | 0.578 | gapdh | -0.207 |
| nkx2.2a | 0.525 | gamt | -0.195 |
| pax6b | 0.509 | ahcy | -0.190 |
| mir7a-1 | 0.504 | hdlbpa | -0.177 |
| isl1 | 0.502 | gatm | -0.174 |
| myt1b | 0.501 | fbp1b | -0.173 |
| si:dkey-153k10.9 | 0.480 | nupr1b | -0.168 |
| pcsk1nl | 0.459 | mat1a | -0.168 |
| scgn | 0.459 | bhmt | -0.167 |
| insm1b | 0.449 | glud1b | -0.163 |
| kcnj11 | 0.445 | cx32.3 | -0.162 |
| lysmd2 | 0.432 | aldh6a1 | -0.161 |
| dlb | 0.423 | eno3 | -0.159 |
| id4 | 0.423 | apoa4b.1 | -0.157 |
| rprmb | 0.417 | aldh7a1 | -0.156 |
| mir375-2 | 0.416 | apoa1b | -0.154 |
| scg2a | 0.401 | gstr | -0.150 |
| syt1a | 0.395 | gstt1a | -0.150 |
| vamp2 | 0.389 | scp2a | -0.149 |
| pygma | 0.388 | si:dkey-16p21.8 | -0.149 |
| LOC100537384 | 0.383 | apoc2 | -0.147 |
| c2cd4a | 0.374 | gnmt | -0.147 |
| gpc1a | 0.369 | agxtb | -0.146 |
| rnasekb | 0.365 | mgst1.2 | -0.146 |
| tspan7b | 0.365 | gstp1 | -0.145 |
| ndufa4l2a | 0.363 | afp4 | -0.143 |
| tox | 0.362 | gcshb | -0.143 |
| cacna1c | 0.359 | aqp12 | -0.142 |
| vat1 | 0.358 | agxta | -0.141 |
| celf3a | 0.358 | fabp3 | -0.140 |
| rem2 | 0.355 | suclg2 | -0.139 |
| tmsb | 0.351 | apoc1 | -0.139 |
| SLC5A10 | 0.350 | aldh1l1 | -0.138 |
| elovl1a | 0.343 | ckba | -0.138 |