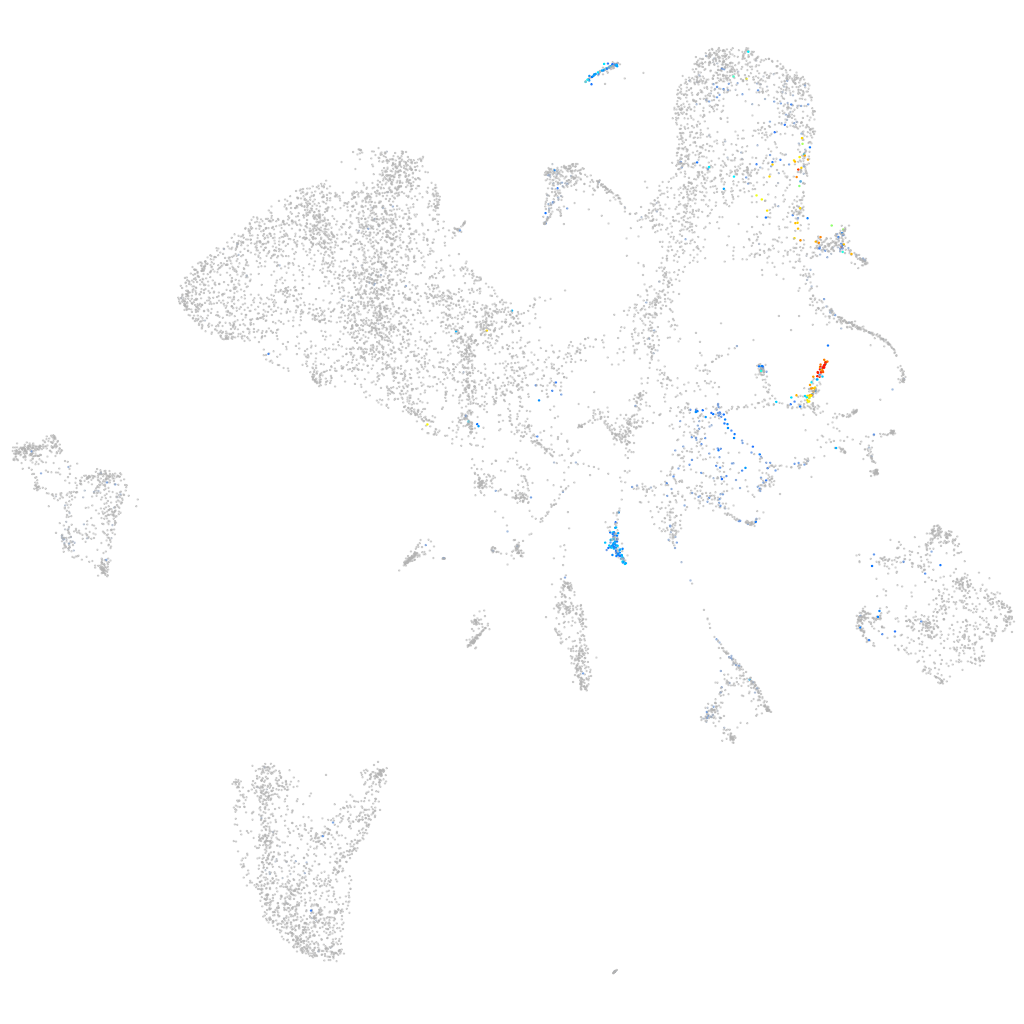
insulin-like 5a
ZFIN 
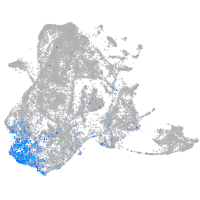
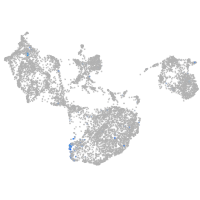
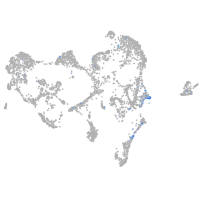
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| galn | 0.542 | fabp3 | -0.149 |
| gcga | 0.401 | ahcy | -0.132 |
| CR556712.1 | 0.391 | bhmt | -0.131 |
| pcsk2 | 0.390 | gamt | -0.128 |
| insl5b | 0.386 | mat1a | -0.122 |
| ccka | 0.383 | aqp12 | -0.121 |
| arxa | 0.370 | gatm | -0.115 |
| calca | 0.344 | apoc1 | -0.115 |
| vipb | 0.338 | agxtb | -0.114 |
| scgn | 0.314 | apoa1b | -0.111 |
| scg3 | 0.313 | kng1 | -0.110 |
| dll4 | 0.312 | tfa | -0.107 |
| egr4 | 0.309 | apoa2 | -0.106 |
| oprd1b | 0.299 | serpinf2b | -0.105 |
| rasd4 | 0.296 | serpina1 | -0.105 |
| LOC101885494 | 0.294 | ttc36 | -0.105 |
| si:ch73-160i9.2 | 0.289 | wu:fj16a03 | -0.105 |
| hepacam2 | 0.281 | grhprb | -0.104 |
| anxa4 | 0.279 | apom | -0.103 |
| gucy2c | 0.277 | rbp2b | -0.103 |
| tac3a | 0.275 | serpina1l | -0.103 |
| c2cd4a | 0.271 | nupr1b | -0.101 |
| adra2a | 0.269 | hao1 | -0.101 |
| rem2 | 0.264 | si:dkey-16p21.8 | -0.100 |
| insm1b | 0.264 | fgg | -0.100 |
| TCIM (1 of many) | 0.261 | ambp | -0.100 |
| slc35g2a | 0.260 | hpda | -0.100 |
| calm1b | 0.259 | hdlbpa | -0.100 |
| alkal2a | 0.258 | aldh6a1 | -0.100 |
| gfra1a | 0.257 | fabp10a | -0.099 |
| pdx1 | 0.253 | f2 | -0.099 |
| fev | 0.241 | agxta | -0.099 |
| tspan34 | 0.239 | zgc:123103 | -0.099 |
| prr15la | 0.236 | uox | -0.099 |
| pfkfb4a | 0.235 | LOC110437731 | -0.099 |