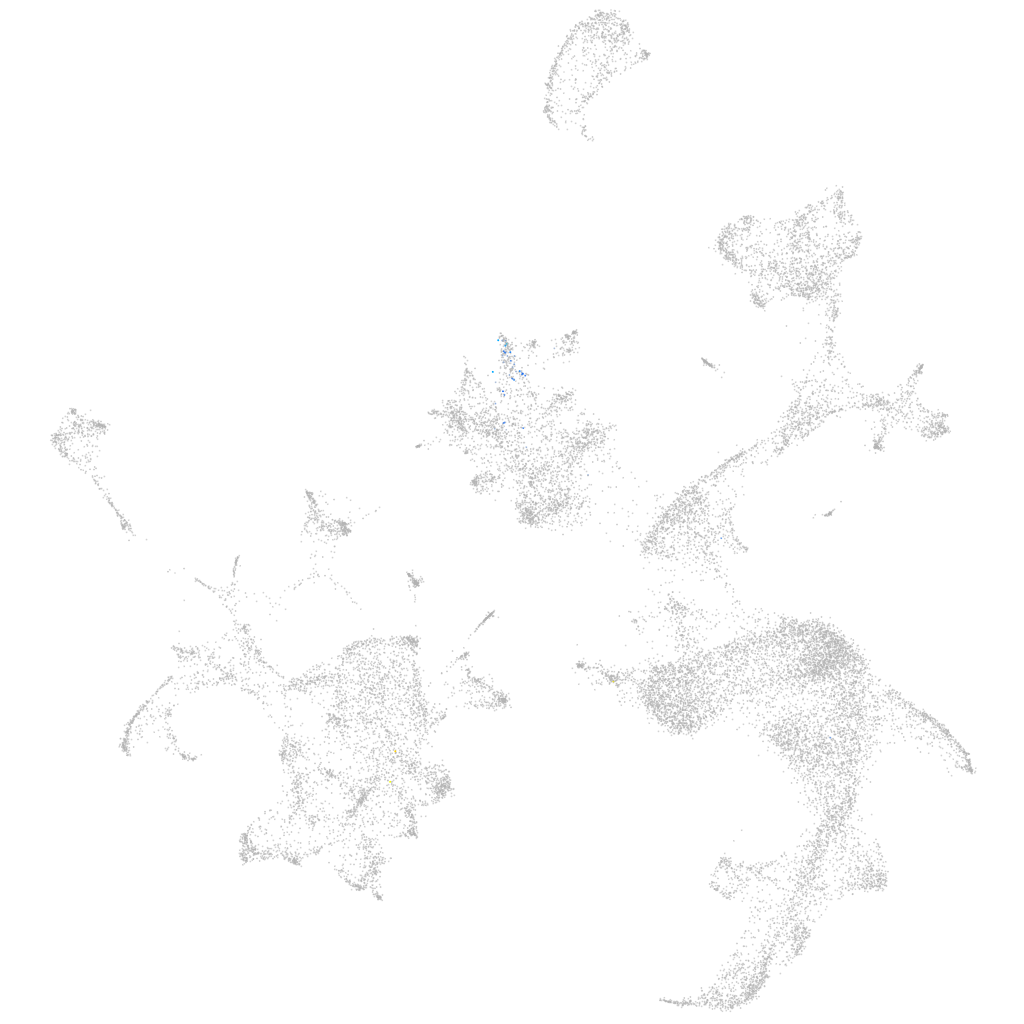
interphotoreceptor matrix proteoglycan 1b
ZFIN 
Other cell groups


all cells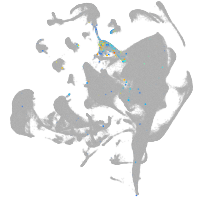 |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 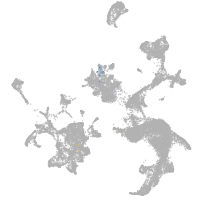 |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| XLOC-019963 | 0.248 | lmo2 | -0.017 |
| cngb3.2 | 0.214 | si:dkey-112e7.2 | -0.016 |
| zgc:103625 | 0.212 | hbbe3 | -0.015 |
| si:ch1073-469d17.2 | 0.211 | arl4aa | -0.014 |
| lrit2 | 0.211 | eif5a2 | -0.014 |
| gabra6a | 0.211 | anp32b | -0.014 |
| tmx3a | 0.207 | clic2 | -0.013 |
| rcvrn2 | 0.202 | etf1b | -0.013 |
| opn6a | 0.202 | abracl | -0.013 |
| guk1b | 0.201 | arhgdig | -0.013 |
| rbp4l | 0.200 | ier2a | -0.013 |
| arl3l1 | 0.199 | znfl2a | -0.013 |
| cnga3a | 0.198 | ptbp1a | -0.013 |
| zgc:73359 | 0.197 | rac2 | -0.012 |
| ckmt2a | 0.197 | pycard | -0.012 |
| inaa | 0.196 | si:dkey-27i16.2 | -0.012 |
| cdhr1a | 0.194 | itm2bb | -0.012 |
| rgs9a | 0.193 | blvra | -0.012 |
| prph2a | 0.193 | urod | -0.012 |
| impg1a | 0.190 | coro1a | -0.012 |
| gnat2 | 0.188 | inka1b | -0.011 |
| atp1b2b | 0.185 | snx5 | -0.011 |
| si:dkeyp-57d7.4 | 0.185 | tmem14ca | -0.011 |
| CR450786.1 | 0.185 | dusp5 | -0.011 |
| prph2b | 0.180 | ywhaz | -0.011 |
| grk7a | 0.178 | dnase1l4.1 | -0.011 |
| tmem237a | 0.177 | arpc1b | -0.011 |
| syt5a | 0.177 | rnaseka | -0.011 |
| si:ch211-285j22.3 | 0.176 | cebpb | -0.011 |
| si:dkey-19c16.12 | 0.176 | zgc:56493 | -0.011 |
| arl3l2 | 0.176 | rhogb | -0.011 |
| elovl4b | 0.176 | mafbb | -0.011 |
| aipl2 | 0.174 | fthl27 | -0.011 |
| grk1b | 0.173 | laptm5 | -0.011 |
| slc25a3a | 0.172 | eif2s2 | -0.011 |