im:7136021
ZFIN 
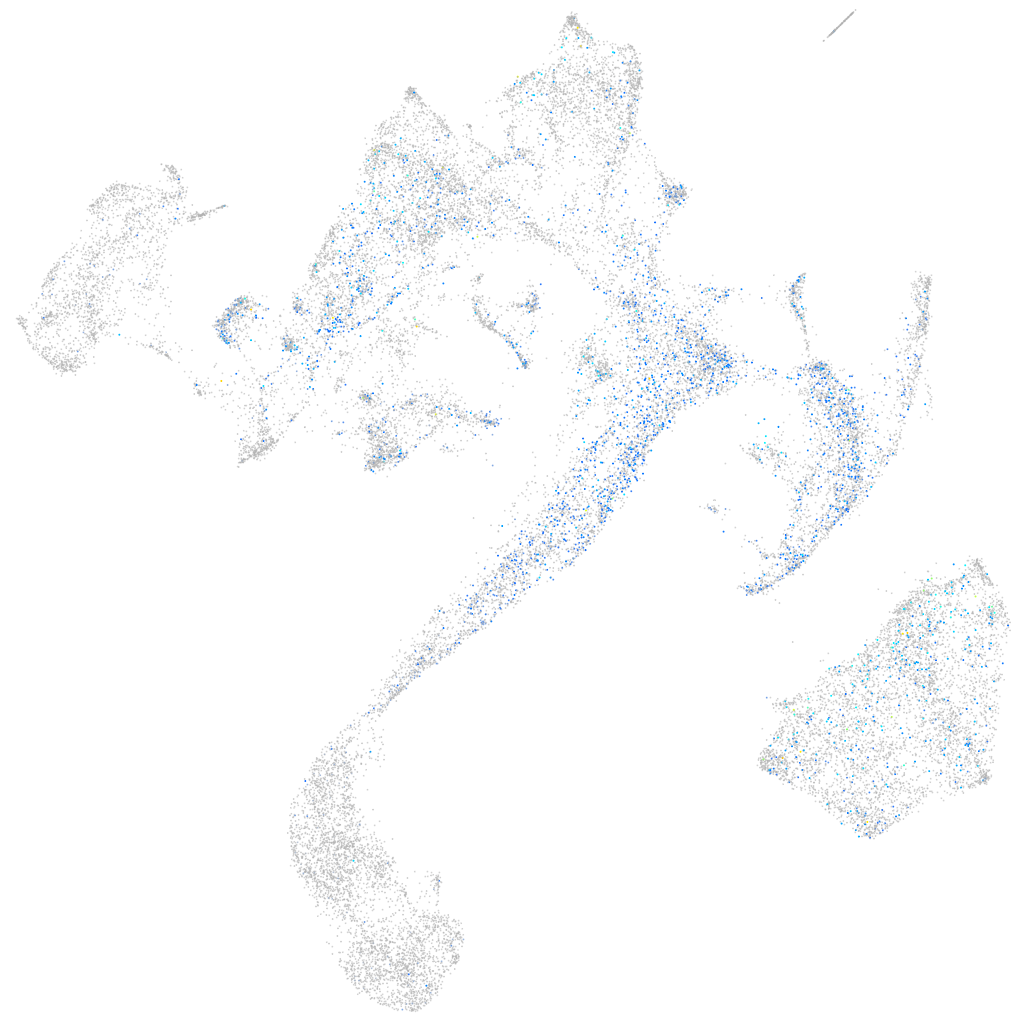
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| snu13b | 0.162 | ckma | -0.120 |
| npm1a | 0.160 | neb | -0.120 |
| setb | 0.154 | tnnc2 | -0.120 |
| rpl7l1 | 0.153 | gapdh | -0.118 |
| nhp2 | 0.150 | actn3a | -0.118 |
| ran | 0.148 | eno3 | -0.117 |
| nop56 | 0.147 | ckmb | -0.117 |
| nop10 | 0.146 | ak1 | -0.117 |
| nop58 | 0.144 | myom1a | -0.116 |
| snrpf | 0.143 | si:ch73-367p23.2 | -0.116 |
| cnbpa | 0.141 | smyd1a | -0.116 |
| nip7 | 0.141 | pgam2 | -0.114 |
| si:dkey-23i12.5 | 0.140 | si:ch211-266g18.10 | -0.113 |
| rcl1 | 0.140 | ldb3b | -0.113 |
| nifk | 0.140 | tmod4 | -0.112 |
| tomm20a | 0.140 | actn3b | -0.112 |
| ssb | 0.139 | si:dkey-16p21.8 | -0.111 |
| dkc1 | 0.138 | atp2a1 | -0.111 |
| snrpd2 | 0.137 | actc1b | -0.111 |
| eif4a1a | 0.136 | casq2 | -0.111 |
| cdkn2aipnl | 0.136 | XLOC-025819 | -0.111 |
| prmt1 | 0.134 | prx | -0.109 |
| mrto4 | 0.133 | aldoab | -0.109 |
| ncl | 0.133 | XLOC-001975 | -0.108 |
| emg1 | 0.133 | acta1b | -0.108 |
| ptges3b | 0.132 | tnnt3a | -0.107 |
| CU929070.1 | 0.132 | ank1a | -0.107 |
| srsf5a | 0.131 | XLOC-005350 | -0.107 |
| bysl | 0.131 | pvalb1 | -0.107 |
| gtpbp4 | 0.131 | nme2b.2 | -0.106 |
| wdr43 | 0.131 | casq1b | -0.105 |
| hnrnpabb | 0.131 | mylpfa | -0.105 |
| ddx21 | 0.130 | pvalb2 | -0.104 |
| naca | 0.129 | cavin4b | -0.104 |
| polr1d | 0.129 | mylz3 | -0.104 |