interleukin enhancer binding factor 3b
ZFIN 
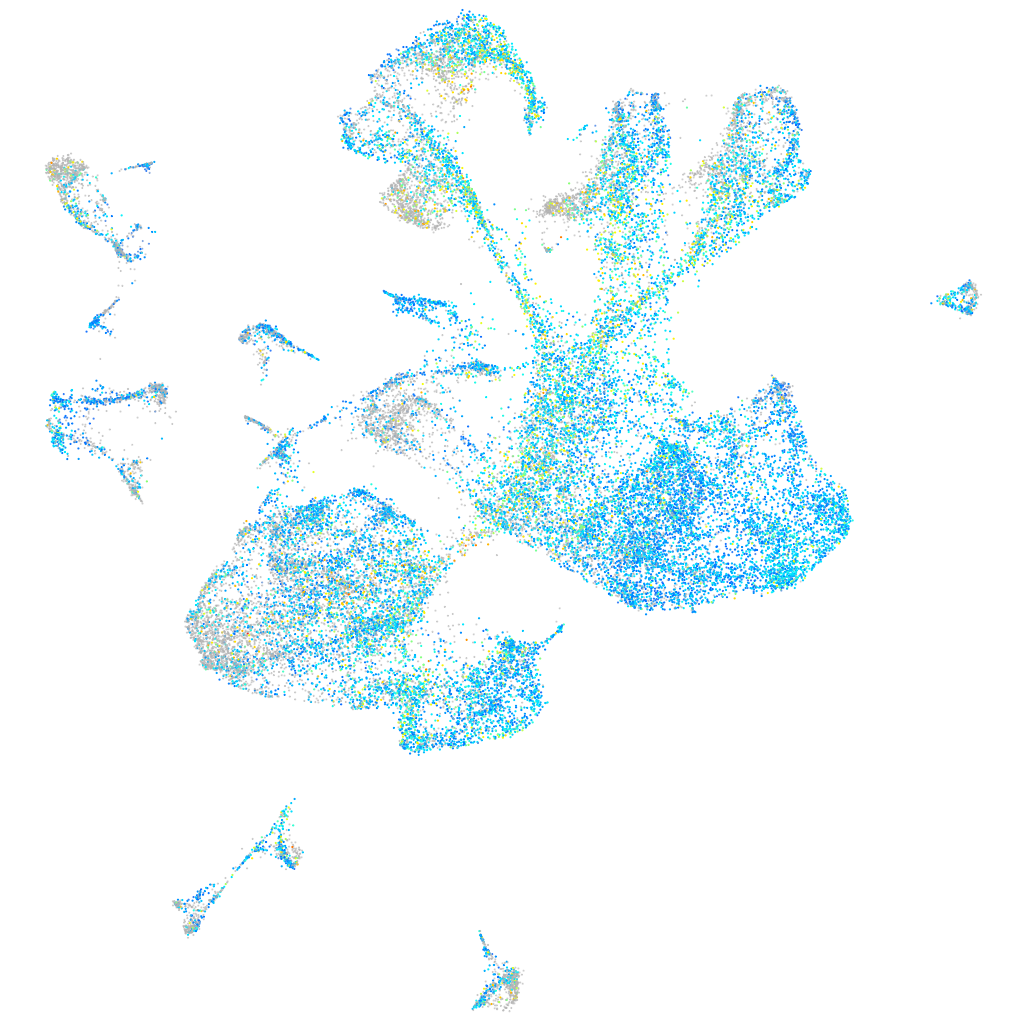
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hnrnpaba | 0.375 | atp1a1b | -0.219 |
| khdrbs1a | 0.374 | glula | -0.212 |
| hnrnpa0l | 0.362 | cx43 | -0.209 |
| hnrnpabb | 0.350 | ptn | -0.199 |
| hmga1a | 0.330 | slc1a2b | -0.196 |
| hnrnpa0b | 0.327 | slc4a4a | -0.190 |
| cirbpb | 0.323 | si:ch211-66e2.5 | -0.186 |
| si:ch211-222l21.1 | 0.323 | slc3a2a | -0.182 |
| ilf2 | 0.314 | acbd7 | -0.176 |
| cirbpa | 0.312 | fabp7a | -0.175 |
| ddx39ab | 0.311 | mt2 | -0.174 |
| snrpd2 | 0.310 | efhd1 | -0.174 |
| hdac1 | 0.309 | cox4i2 | -0.174 |
| h2afvb | 0.306 | hepacama | -0.173 |
| ptmab | 0.303 | CR383676.1 | -0.173 |
| snrpf | 0.299 | zgc:153704 | -0.169 |
| alyref | 0.293 | ppap2d | -0.168 |
| hmgb2b | 0.291 | cebpd | -0.165 |
| snrpg | 0.291 | gpr37l1b | -0.164 |
| tubb2b | 0.290 | apoa2 | -0.163 |
| syncrip | 0.288 | qki2 | -0.163 |
| seta | 0.288 | mdkb | -0.160 |
| si:ch211-288g17.3 | 0.287 | cldn7a | -0.155 |
| hspa8 | 0.283 | ndrg3a | -0.154 |
| sumo3a | 0.279 | cdo1 | -0.150 |
| ran | 0.279 | pvalb1 | -0.150 |
| h3f3a | 0.278 | cyp2ad3 | -0.150 |
| psmb7 | 0.278 | smox | -0.150 |
| cct5 | 0.277 | gapdhs | -0.149 |
| eef1a1l1 | 0.276 | dap1b | -0.148 |
| apex1 | 0.275 | mfge8a | -0.147 |
| snrpe | 0.274 | lpin1 | -0.147 |
| nono | 0.274 | apoa1b | -0.145 |
| marcksb | 0.274 | anxa13 | -0.145 |
| hsp90ab1 | 0.273 | IGLON5 | -0.144 |