interleukin 6 receptor
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 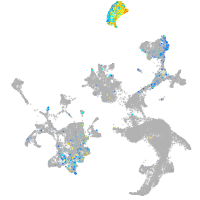 |
pronephros 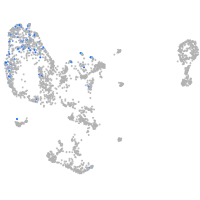 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line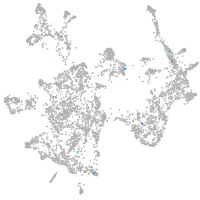 |
epidermis |
periderm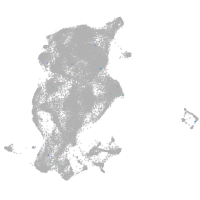 |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC103911520 | 0.489 | ptmab | -0.082 |
| crygm2d18 | 0.426 | sec61b | -0.078 |
| crygm2d13 | 0.407 | tma7 | -0.069 |
| crybb1l1 | 0.373 | hmga1a | -0.068 |
| fgf6a | 0.370 | hnrnpaba | -0.068 |
| LOC101886038 | 0.368 | hsp90ab1 | -0.067 |
| fam102aa | 0.354 | hnrnpa0b | -0.067 |
| LOC110439359 | 0.333 | setb | -0.062 |
| XLOC-001835 | 0.329 | tbxta | -0.061 |
| fam228a | 0.321 | hnrnpabb | -0.060 |
| CU929365.1 | 0.303 | ssr2 | -0.059 |
| LOC110438016 | 0.294 | snrpg | -0.059 |
| LOC101882135 | 0.289 | dad1 | -0.059 |
| dram2a | 0.289 | snrpb | -0.058 |
| mc1r | 0.285 | h2afvb | -0.058 |
| crygm2d10 | 0.284 | snrpf | -0.058 |
| pcdh20 | 0.283 | rpl4 | -0.058 |
| crygm2d3 | 0.273 | msna | -0.057 |
| crygmx | 0.263 | selenof | -0.057 |
| cntnap3 | 0.253 | ybx1 | -0.057 |
| plac8l1 | 0.253 | snrpe | -0.057 |
| crygm2d8 | 0.240 | snrpd2 | -0.057 |
| LOC103910903 | 0.239 | h2afva | -0.057 |
| CABZ01084230.1 | 0.238 | snrpd1 | -0.056 |
| grik4 | 0.237 | serbp1a | -0.055 |
| znf804a | 0.232 | qkia | -0.055 |
| slc30a10 | 0.229 | hspa8 | -0.055 |
| epdl1 | 0.223 | eif4a1a | -0.054 |
| vill | 0.222 | calm2b | -0.054 |
| BX000438.2 | 0.222 | pabpc1a | -0.053 |
| prg4b | 0.220 | cirbpa | -0.053 |
| ptgis | 0.219 | atp5mc1 | -0.053 |
| stard15 | 0.217 | h3f3d | -0.053 |
| si:ch211-190k17.19 | 0.216 | tubb2b | -0.053 |
| crygs1 | 0.216 | hspa5 | -0.053 |