interleukin 17 receptor D
ZFIN 
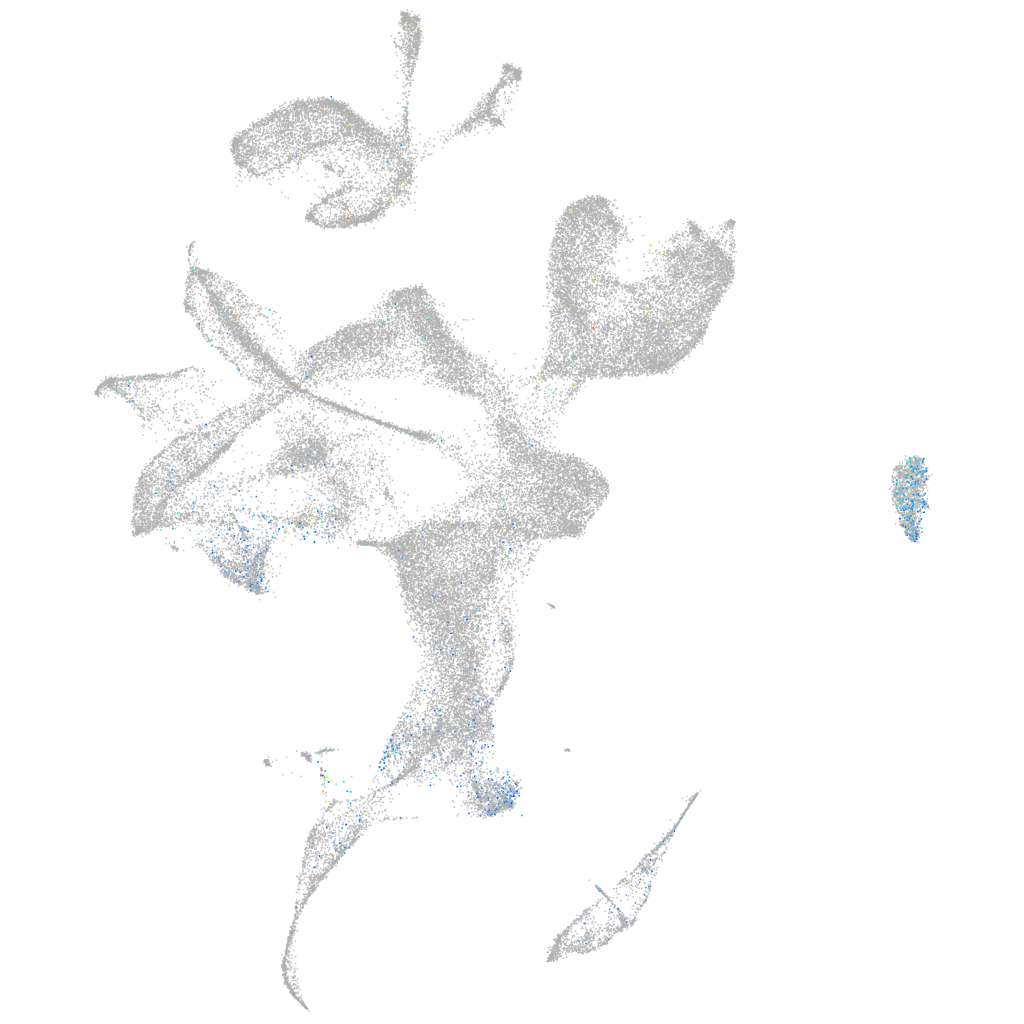
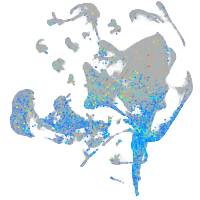
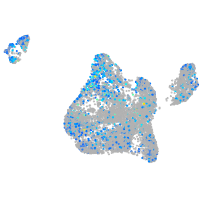
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hspb1 | 0.188 | ptmaa | -0.082 |
| apoeb | 0.157 | h3f3a | -0.081 |
| sox19a | 0.152 | ppiab | -0.071 |
| si:ch211-152c2.3 | 0.151 | rpl37 | -0.067 |
| stm | 0.151 | ckbb | -0.067 |
| shisa2a | 0.150 | cadm3 | -0.066 |
| aldob | 0.149 | si:ch1073-429i10.3.1 | -0.065 |
| COX7A2 | 0.145 | gpm6aa | -0.064 |
| tdgf1 | 0.137 | tuba1c | -0.062 |
| nr6a1a | 0.137 | rps10 | -0.061 |
| pax2a | 0.135 | gpm6ab | -0.060 |
| fgf3 | 0.135 | fabp7a | -0.058 |
| pprc1 | 0.133 | foxg1b | -0.057 |
| hesx1 | 0.131 | CR383676.1 | -0.057 |
| pkdccb | 0.131 | zgc:114188 | -0.055 |
| zmp:0000000624 | 0.130 | actc1b | -0.053 |
| npm1a | 0.126 | pvalb1 | -0.051 |
| cyp26a1 | 0.126 | hnrnpa0l | -0.051 |
| nop2 | 0.124 | epb41a | -0.050 |
| s100a1 | 0.124 | rnasekb | -0.050 |
| fbl | 0.122 | gapdhs | -0.050 |
| bms1 | 0.122 | COX3 | -0.050 |
| dkc1 | 0.121 | h3f3c | -0.048 |
| gnl3 | 0.121 | btg1 | -0.048 |
| nop58 | 0.119 | atp6v0cb | -0.047 |
| chrd | 0.119 | atp6v1g1 | -0.045 |
| alcamb | 0.118 | zgc:153409 | -0.045 |
| NC-002333.4 | 0.118 | hbbe1.3 | -0.045 |
| cxcl12a | 0.118 | ptmab | -0.044 |
| apoc1 | 0.117 | snap25b | -0.044 |
| gar1 | 0.116 | mt-co2 | -0.044 |
| si:ch211-212k18.5 | 0.116 | pvalb2 | -0.043 |
| sfrp1a | 0.115 | neurod4 | -0.043 |
| apela | 0.114 | hbae3 | -0.043 |
| rsl1d1 | 0.114 | rorb | -0.042 |