"immunoglobulin superfamily, member 9b"
ZFIN 
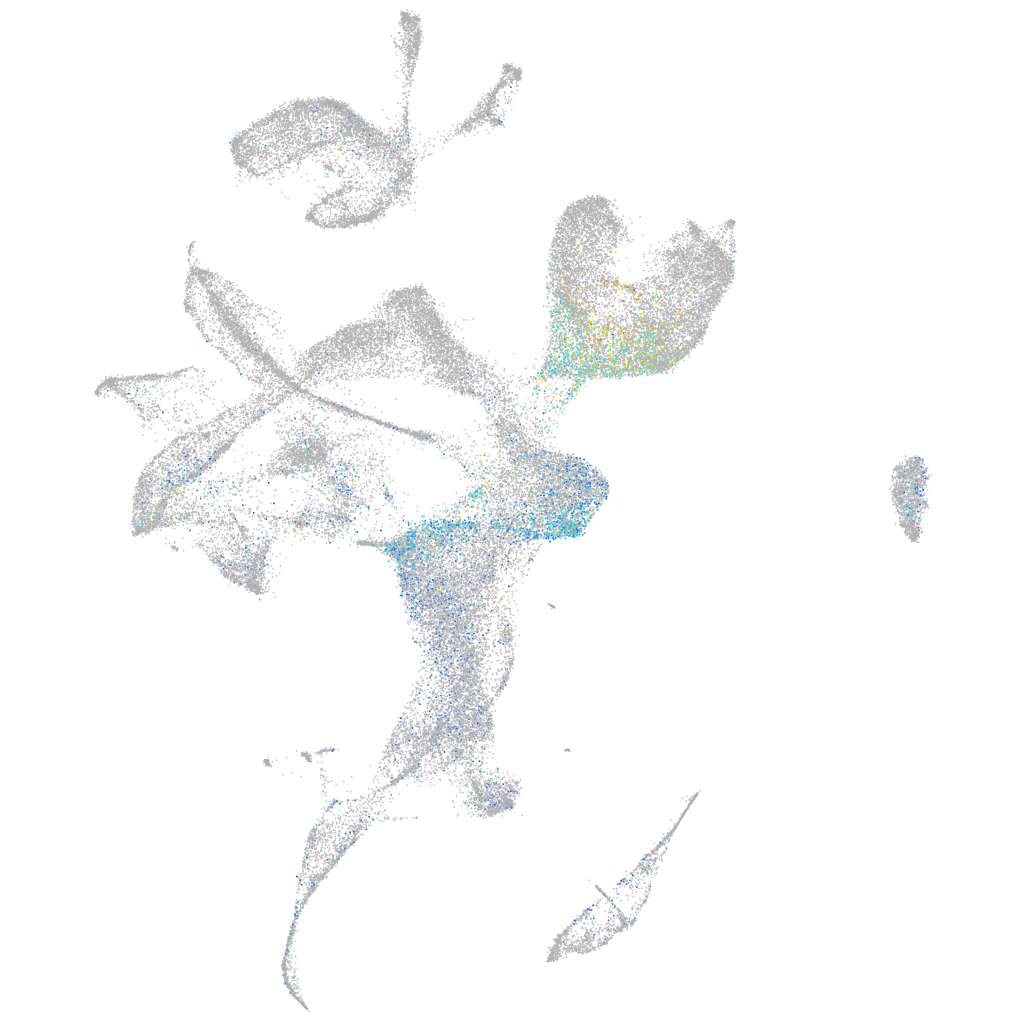
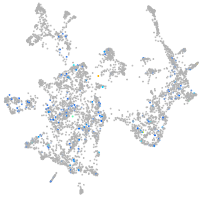
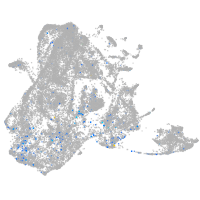
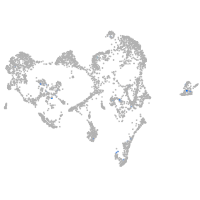
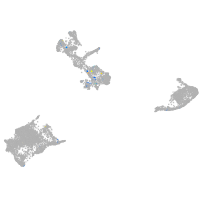
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| vsx1 | 0.156 | gapdhs | -0.091 |
| ptprua | 0.154 | atp5mc1 | -0.086 |
| scrt2 | 0.154 | sncb | -0.074 |
| rorb | 0.152 | dynll1 | -0.071 |
| hunk | 0.152 | c1qbp | -0.070 |
| btg1 | 0.152 | cox6a1 | -0.070 |
| tp53inp1 | 0.149 | elavl3 | -0.069 |
| bcl2l10 | 0.145 | pax6b | -0.068 |
| ndrg1b | 0.144 | pax6a | -0.068 |
| myt1b | 0.142 | eno1a | -0.067 |
| mdkb | 0.131 | atp5pf | -0.067 |
| otx2b | 0.130 | cox7a2a | -0.067 |
| neurod4 | 0.129 | gpx4b | -0.067 |
| nol4la | 0.127 | si:dkey-16p21.8 | -0.066 |
| im:7152348 | 0.124 | mab21l1 | -0.066 |
| sept4a | 0.123 | cox8a | -0.066 |
| ccdc88c | 0.122 | ptges3a | -0.066 |
| tp53inp2 | 0.121 | sncgb | -0.066 |
| ddah2 | 0.118 | gng3 | -0.065 |
| CABZ01055522.1 | 0.116 | atp5mf | -0.064 |
| prickle1b | 0.116 | uqcrh | -0.063 |
| CU634008.1 | 0.116 | hsp90aa1.2 | -0.063 |
| nova2 | 0.116 | sh3gl2a | -0.063 |
| cirbpb | 0.115 | atp6v0cb | -0.062 |
| olig2 | 0.115 | COX5B | -0.062 |
| kdm2bb | 0.115 | hspe1 | -0.062 |
| cadm3 | 0.115 | aldoaa | -0.061 |
| baz2ba | 0.114 | atp5meb | -0.060 |
| smad7 | 0.112 | hmgn7 | -0.060 |
| hmgb1a | 0.112 | cox7c | -0.060 |
| marcksl1a | 0.109 | snap25b | -0.059 |
| fkbp1ab | 0.108 | mir181b-3 | -0.059 |
| ankrd12 | 0.108 | cox7b | -0.058 |
| sox11a | 0.105 | nop10 | -0.058 |
| gpm6aa | 0.105 | chchd10 | -0.058 |