"immunoglobulin superfamily, member 3"
ZFIN 
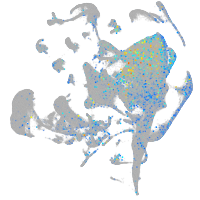
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| myt1b | 0.097 | si:ch211-66e2.5 | -0.059 |
| elavl3 | 0.094 | atp1a1b | -0.058 |
| nova2 | 0.081 | cx43 | -0.055 |
| elavl4 | 0.080 | id1 | -0.054 |
| rtn1a | 0.078 | glula | -0.052 |
| rtn1b | 0.077 | slc4a4a | -0.051 |
| cplx2 | 0.075 | ptn | -0.050 |
| tubb5 | 0.074 | fosab | -0.049 |
| syt2a | 0.074 | sparc | -0.049 |
| hmgb3a | 0.073 | ppap2d | -0.049 |
| ywhah | 0.073 | vamp3 | -0.048 |
| nova1 | 0.073 | cd63 | -0.047 |
| golga7ba | 0.073 | ptgdsb.2 | -0.047 |
| stx1b | 0.073 | slc3a2a | -0.046 |
| stxbp1a | 0.071 | efhd1 | -0.046 |
| gdf11 | 0.071 | cox4i2 | -0.045 |
| tmsb | 0.070 | her9 | -0.044 |
| dpysl3 | 0.069 | slc1a3b | -0.044 |
| tmem59l | 0.069 | fabp7a | -0.043 |
| thsd7aa | 0.067 | cebpd | -0.043 |
| si:dkey-276j7.1 | 0.067 | qki2 | -0.043 |
| atp1b2a | 0.067 | pgrmc1 | -0.043 |
| tuba2 | 0.067 | mt2 | -0.042 |
| sox11b | 0.067 | selenoh | -0.042 |
| sncb | 0.066 | tpm4a | -0.042 |
| tuba1c | 0.066 | si:ch211-286b5.5 | -0.042 |
| stmn1b | 0.066 | hepacama | -0.041 |
| gng2 | 0.065 | GCA | -0.041 |
| vamp2 | 0.065 | ptgdsb.1 | -0.041 |
| baz2ba | 0.065 | zgc:153704 | -0.041 |
| rbfox1 | 0.064 | npm1a | -0.041 |
| vsnl1b | 0.064 | si:dkey-151g10.6 | -0.041 |
| ndrg4 | 0.064 | psat1 | -0.041 |
| mab21l2 | 0.064 | mcm2 | -0.040 |
| myt1a | 0.064 | sept8b | -0.040 |